TP5 - SyMBoD
Integrative system medicine platform
System medicine platforms require the integration of clinical and research data, mathematical and computational models and user-friendly tools for efficient, personalized diagnoses and identification of therapy options („theranostics“). For SyMBoD we develop a digital theranostics platform to enable personalized regenerative therapy options for patients with critical bone defects. To make our research data findable, accessible, interoperable and reusable (FAIR) and to meet the needs of platform users, storage, access, manipulation and distribution of the data generated from the consortium is facilitated by intuitive user interfaces. In particular, we developed a framework for semantic integration of clinical and multi-omics data from the other subprojects and external knowledge resources by harmonizing metadata across several data sources and providing annotation and integration tools for experimentalists. A corresponding platform prototype for the consortium partners has been deployed during the first funding period on de.NBI resources.
Furthermore, we are currently integrating into this platform exploratory and predictive bioinformatics and machine learning analyses for optimization of simulations and models as well as for stratification of patients. Here, we build upon existing and novel bioinformatics methods for network-based identification of system biology driven biomarkers and de novo stratification of patients into mechanistic endophenotypes. Phenotypic parameters obtained this way will be directly feedable into CAD/CAE simulations and models developed in SP4, which have been successfully integrated in their independent form as interactive workflows within the platform.
Rich interactive visualizations for hypothesis generation based on integrative data analyses and therapy option suggestion using individually optimized 3D scaffold models will allow platform users to perform end-to-end individualized bone regenerative therapy optimizations. Ultimately, we will advance the research platform into a patient-centric system for personalized bone regenerative therapy that is optimized towards clinicians’ needs and practices and evaluate opportunities for its commercial exploitation. This subproject is augmented by the collaboration between the Baumbach Lab at UHH and Genevention GmbH who have extensive software development experience and state-of-the-art scientific expertise in the field.
Clinical and experimental omics data are linked with each other in the SyMBoD theranostics platform and are sustainably available, including their rich annotation and references to external knowledge resources.
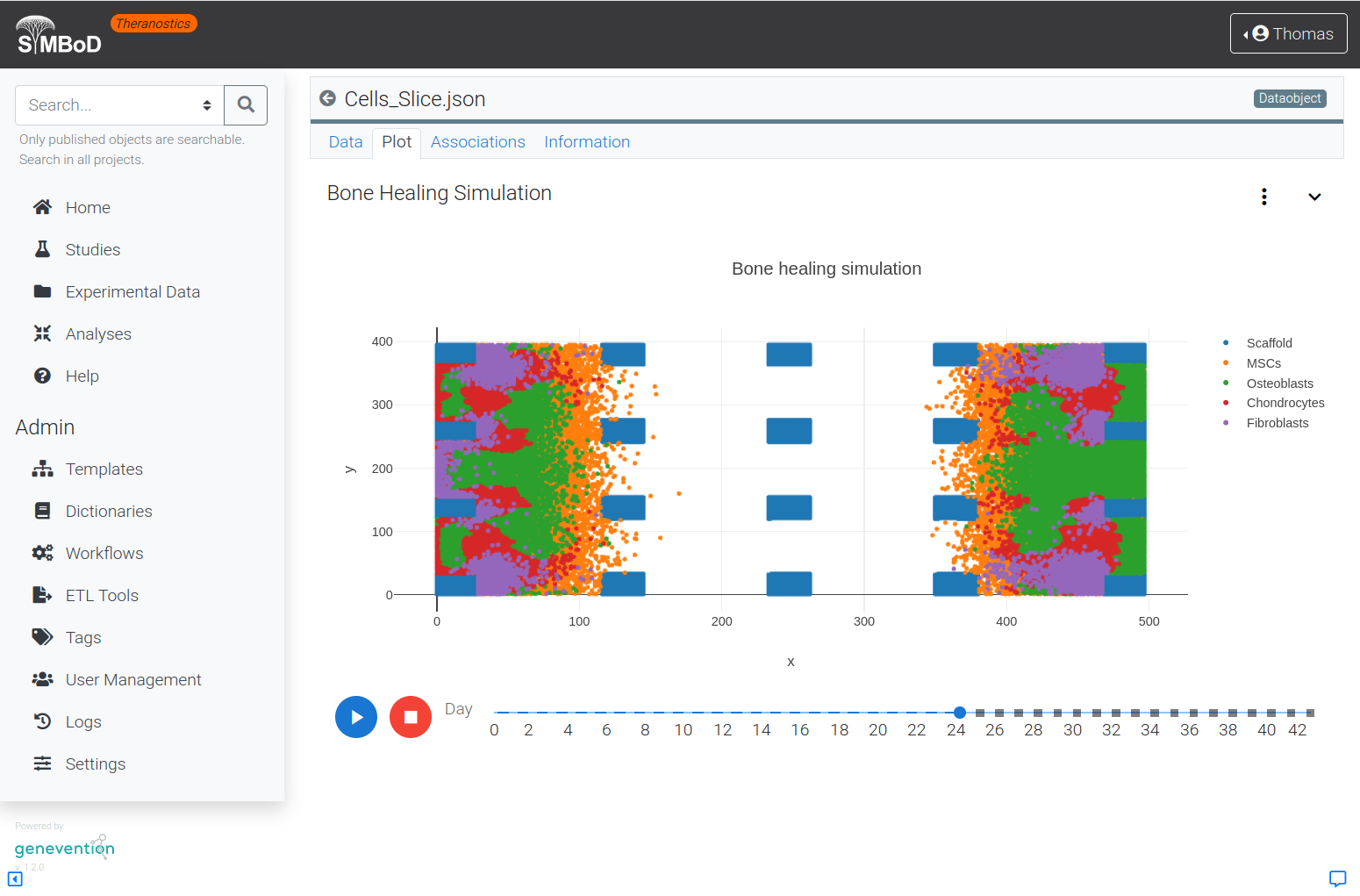
The SyMBoD theranostics platform allows simulation of cell differentiation and proliferation during regeneration of bone tissue around a 3D-printed scaffold made of resorbable material by means of easy-to-use user interfaces.



