SP 2
Image analysis of cytological TMM-features
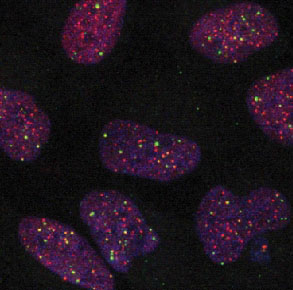
The task of subproject SP2 is the development and application of image analysis methods for automatic extraction of cytological features from 3D multi-channel microscopy images. The focus is on the determination of features, which are relevant for Telomere Maintenance Mechanisms (TMMs). We develop a model-based image analysis approach for 3D localization an colocalization of subcellular structures, and apply it to 3D confocal microscopy images of different cell lines (e.g., glioblastoma) as well as to 3D microscopy images of tissue microarrays (TMAs).

From the image data a wealth of quantitative information on cytological features of different TMMs can be extracted. This includes, for example, telomere length distribution and heterogeneity for classification into ALT (Alternative Lengthening of Telomeres) positive or negative cells, the average telomere length, as well as the number and size of APBs (ALT associated PML bodies) as a hallmark of ALT by colocalization of telomeres and PML proteins. The automatically extracted cytological TMM features are important for the modeling of TMM networks in tumor by an integrativesystems biology approach as well as for stratification of tumor entities.
Keywords: Computer-based image analysis, cytological features, 3D multi-channel microscopy images, tissue microarrays, telomere maintenance mechanism, alternative lengthening of telomeres, 3D colocalization



