SP 4
SNP-mediated miRNA (dys)regulation in atherosclerosis
The involvement of microRNAs (small non-coding RNAs) in atherosclerosis is still poorly understood, yet there is some evidence that genetic variance (single nucleotide polymorphisms, SNPs) may compromise the expression or activity of microRNAs and thereby engage atherosclerotic processes. Our joint subproject 4 follows two key hypotheses which will be addressed in seven work packages: 1) SNPs in microRNA genes or other genes (e.g. for transcription factors) alter microRNA expression or 2) SNPs in microRNA binding sites disturb microRNA control over certain mRNAs (see Fig.).
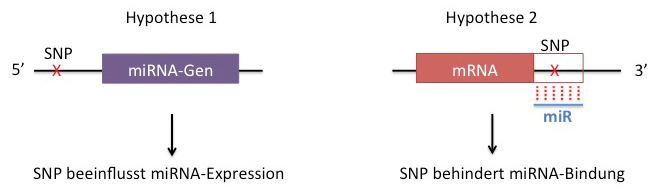
In pursuit of these hypotheses, we will comprehensively identify such loci of genetic variance (termed expressed quantitaive trait loci, eQTL) and characterize their impact with respect to disease. First, we will quantifiy 1,700 microRNAs in samples from 150 patients with diagnosed coronary artery disease (CAD) or stroke (WP4.1) and then correlate these eQTLs of microRNAs with disease in the patients collective (WP4.2). To validate these findings experimentally, we will test by state-of-the-art microRNA methodology and fluorescence microscopy whether experimentally altered microRNA expression/activity increases proliferation of vascular smooth muscle cells (VSMCs) - a hallmark of developing atherosclerosis (WP4.3). As for hypothesis 2, we will search genotype datasets from CAD/stroke patients for the existence of SNPs in microRNA binding sites of atherosclerosis genes (WP4.4). Whether these SNPs truly impair the ability of a microRNA to control its taget mRNA(s) will be studied in luciferase reporter assays (WP4.5). In parallel, we will develop an in silico analysis method (cross-OMICs enrichment) to clarify whether newly identified microRNA eQTLs comply with known atherosclerosis pathways or to subselect new pathways WP4.6). We use a Bayesian framework for pathway analyses, since it allows the use of microRNA sets instead of standard gene set analysis and we aim to extend the analysis beyond a single molecular species. Finally, we plan to validate in vivo by genetic deletion of two examplary, SNP-affected target genes whether these contribute to atherosclerosis (WP4.7).
Keywords: SNP, microRNA, eQTL



