Quan-T-cell
Quantitative T cell immunology to inform immunotherapy and vaccination
Aims:
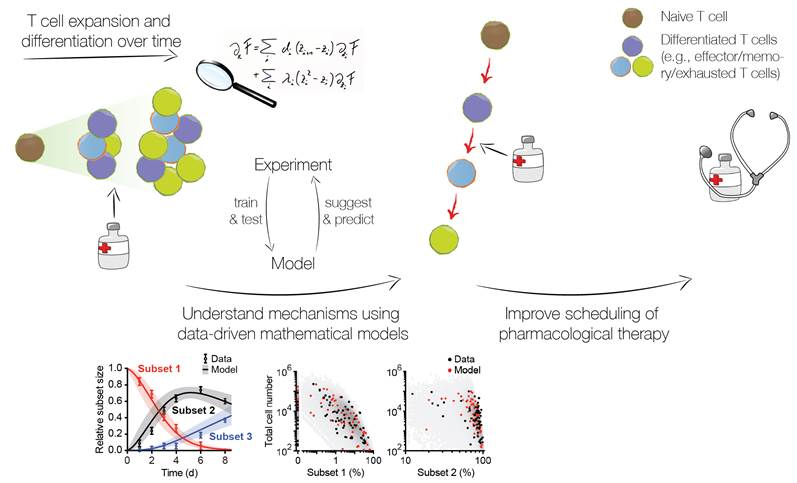
Adoptive T cell therapy (ACT) has recently shown very encouraging results in the treatment of cancer and chronic viral infections. In parallel, pharmacological therapies using immune checkpoint blockers that reverse a state of T cell dysfunction termed "exhaustion" are emerging as a promising treatment for even advanced cancers. However, the outcome of these immunotherapies is largely unpredictable by current clinical parameters, and remarkable therapy success in one patient (e.g., cure from advanced melanoma) still contrasts with failure in another. Thus, there is an urgent need for optimizing therapy and defining prospective parameters for patient stratification. T cells differentiate during immune responses into different kinds of effector and memory subsets with distinct functions. However, the differentiation pathways of memory, effector and exhausted T cells, their developmental plasticity, and possibilities for molecular intervention remain poorly understood.
We have previously shown that data-driven mathematical modeling combined with single-T-cell fate-mapping provides a novel, highly informative approach to study T cell differentiation and plasticity in vivo.
In this collaborative project we will utilize this combined strength of theory and in vivo fate-mapping and
- (1) dissect the mechanisms of T cell differentiation and exhaustion and
- (2) apply this knowledge to optimize ACT and pharmacological therapy as well as vaccination strategies.
Methods:
Based on mouse models of chronic viral infection and melanoma, we will develop mathematical models of T-cell memory formation and exhaustion to identify the as yet unknown dynamic relationship between terminally exhausted and therapeutically valuable T cell subsets. We will infer differentiation pathways from data, elucidate underlying molecular networks and define the role of antigen avidity as a potential key quantitative parameter for T cell fate and therapeutic potential. These mechanistic insights will be applied to find optimal antigen-specific T cell subsets for ACT and improve the scheduling of pharmacological therapy.
Publications
Cho, Y.-L., M. Flossdorf, L. Kretschmer, T. Höfer, D. H. Busch, and V. R. Buchholz (2017). "TCR Signal Quality Modulates Fate Decisions of Single CD4(+) T Cells in a Probabilistic Manner." Cell Reports 20(4): 806-818. Quan-T-Cell doi.org/10.1016/j.celrep.2017.07.005.
Flommersfeld, S., J. P. Bottcher, J. Ersching, M. Flossdorf, P. Meiser, L. O. Pachmayr, J. Leube, I. Hensel, S. Jarosch, Q. Zhang, M. Z. Chaudhry, I. Andrae, M. Schiemann, D. H. Busch, L. Cicin-Sain, J. C. Sun, G. Gasteiger, G. D. Victora, T. Hofer, V. R. Buchholz, and S. Grassmann (2021). "Fate mapping of single NK cells identifies a type 1 innate lymphoid-like lineage that bridges innate and adaptive recognition of viral infection." Immunity 54(10): 2288-2304 e2287. Quan-T-cell www.ncbi.nlm.nih.gov/pubmed/34437840.
Grassmann, S., L. Mihatsch, J. Mir, A. Kazeroonian, R. Rahimi, S. Flommersfeld, K. Schober, I. Hensel, J. Leube, L. O. Pachmayr, L. Kretschmer, Q. Zhang, A. Jolly, M. Z. Chaudhry, M. Schiemann, L. Cicin-Sain, T. Höfer, D. H. Busch, M. Flossdorf, and V. R. Buchholz (2020). "Early emergence of T central memory precursors programs clonal dominance during chronic viral infection." Nat Immunol 21(12): 1563–1573. Quan-T-cell doi.org/10.1038/s41590-020-00807-y.
Kretschmer, L., M. Flossdorf, J. Mir, Y.-L. Cho, M. Plambeck, I. Treise, A. Toska, S. Heinzel, M. Schiemann, D. H. Busch, and V. R. Buchholz (2020). "Differential expansion of T central memory precursor and effector subsets is regulated by division speed." Nat. Commun. 11(113): 1–12. Quan-T-Cell doi.org/10.1038/s41467-019-13788-w.
Pinto-Sietsma, S.-J., M. Flossdorf, V. R. Buchholz, J. Offerhaus, H. Bleijendaal, M. Beudel, P. G. A. Volders, R. M. A. ter Bekke, T. Dormans, P.-P. Zwetsloot, P. de Jager, S. Massberg, P. Rämer, C. Wendtner, E. Hoffmann, K. Rothe, S. Feihl, T. Kessler, Y. M. Pinto, and H. Schunkert (2020). "Antihypertensive drugs in COVID-19 infection." Eur Heart J Cardiovasc Pharmacother. AbCD-Net & Quan-T-cell doi.org/10.1093/ehjcvp/pvaa058.
Plambeck, M., A. Kazeroonian, D. Loeffler, L. Kretschmer, C. Salinno, T. Schroeder, D. H. Busch, M. Flossdorf, and V. R. Buchholz (2022). "Heritable changes in division speed accompany the diversification of single T cell fate." Proc Natl Acad Sci U S A 119(9). Quan-T-cell www.ncbi.nlm.nih.gov/pubmed/35217611.
Poschke, I., M. Flossdorf, and R. Offringa (2016). "Next-generation TCR sequencing - a tool to understand T-cell infiltration in human cancers." The Journal of Pathology 240(4): 384-386. Quan-T-cell doi.org/10.1002/path.4800.



