ComorbSysMed
Comorbidity patterns in inflammatory skin diseases: A systems medicine approach using machine learning and omics technologies
Aims:
Many patients with chronic inflammatory skin diseases, including atopic dermatitis and psoriasis, develop auto-immune mediated comorbidities affecting other barrier organs. However, the underlying pathophysiological and molecular mechanisms are largely unknown. The Junior Research Group will follow a systems medicine approach to improve our understanding of these cross-phenotype aetiologies, integrating multiple levels of omics data, clinical information and external biological knowledge in mathematical models that allow stratification of patients according to their comorbidity status. The main focus will be on the interpretability of the created models thereby enabling the in silico identification of key molecules, pathways and biological networks.
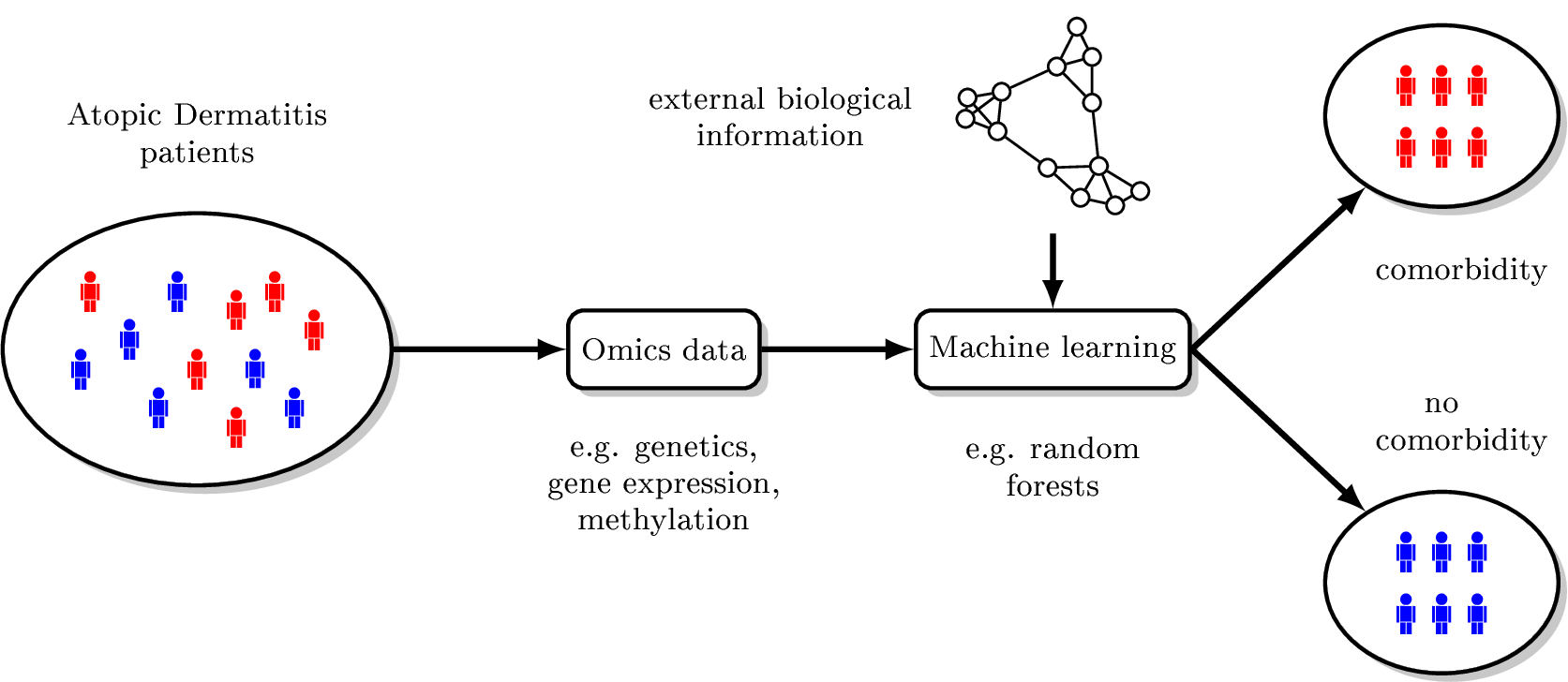
Workplan:
Machine-learning techniques will be employed to tackle the specific challenges of analysing high-dimensional omics data sets. Novel approaches will be developed to enrich the set of possible molecular risk factors for those that are relevant for classification, to incorporate information about structural and functional relationships between molecules into the model training step and to build an integrated model based on multiple levels of omics data. Extensive simulation studies will be performed to understand strengths and weaknesses of both novel and existing machine learning methods. Both, methods and the simulation framework, will be implemented as packages of the freely available and widely used statistical software R. Several patient collections with different types of omics data measured in whole blood or skin biopsies are available to the group. In collaboration with clinical partners, these collections will be extended further by specifically recruiting patients with selected comorbidities.
Publications
Degenhardt, F., S. Seifert, and S. Szymczak (2017). "Evaluation of variable selection methods for random forests and omics data sets." Briefings in Bioinformatics. ComorbSysMed doi.org/10.1093/bib/bbx124.
Gundlach, S., O. Junge, L. Wienbrandt, M. Krawczak, and A. Caliebe (2019). "Comparison of Markov Chain Monte Carlo Software for the Evolutionary Analysis of Y-Chromosomal Microsatellite Data." Comput Struct Biotechnol J 17: 1082-1090. CoMorbSysMed doi.org/10.1016/j.csbj.2019.07.014.
Hu, J., and S. Szymczak (2023). "A review on longitudinal data analysis with random forest." Briefings Bioinf 24(2). CoMorbSysMed doi.org/10.1093/bib/bbad002.
Kachroo, P., S. Szymczak, F.-A. Heinsen, M. Forster, J. Bethune, G. Hemmrich-Stanisak, L. Baker, M. Schrappe, M. Stanulla, and A. Franke (2018). "NGS-based methylation profiling differentiates TCF3-HLF and TCF3-PBX1 positive B-cell acute lymphoblastic leukemia." Epigenomics 10(2): 133-147. ComorbSysMed & SysINFLAME doi.org/10.2217/epi-2017-0080.
Schlicht, K., P. Nyczka, A. Caliebe, S. Freitag-Wolf, A. Claringbould, L. Franke, U. Vosa, B. Consortium, S. L. R. Kardia, J. A. Smith, W. Zhao, C. Gieger, A. Peters, H. Prokisch, K. Strauch, K. S. Group, H. Baurecht, S. Weidinger, P. Rosenstiel, M. T. Hutt, C. Knecht, S. Szymczak, and M. Krawczak (2019). "The metabolic network coherence of human transcriptomes is associated with genetic variation at the cadherin 18 locus." Hum Genet. SysINFLAME & ComorbSysMed www.ncbi.nlm.nih.gov/pubmed/30852652.
Seifert, S. (2020). "Application of random forest based approaches to surface-enhanced Raman scattering data." Sci Rep 10(5436): 1–11. CoMorbSysMed doi.org/10.1038/s41598-020-62338-8.
Seifert, S., S. Gundlach, O. Junge, and S. Szymczak (2020). "Integrating biological knowledge and gene expression data using pathway guided random forests: A benchmarking study." Bioinformatics. ComorbSysMed & coNfirm https://doi.org/10.1093/bioinformatics/btaa483.
Seifert, S., S. Gundlach, and S. Szymczak (2019). "Surrogate minimal depth as an importance measure for variables in random forests." Bioinformatics 35(19): 3663-3671. ComorbSysMed doi.org/10.1093/bioinformatics/btz149.
Tsoi, L. C., E. Rodriguez, F. Degenhardt, H. Baurecht, U. Wehkamp, N. Volks, S. Szymczak, W. R. Swindell, M. K. Sarkar, K. Raja, S. Shao, M. Patrick, Y. Gao, R. Uppala, B. E. Perez White, S. Getsios, P. W. Harms, E. Maverakis, J. T. Elder, A. Franke, J. E. Gudjonsson, and S. Weidinger (2019). "Atopic dermatitis is an IL-13 dominant disease with greater molecular heterogeneity compared to psoriasis." J Invest Dermatol. ComorbSysMed doi.org/10.1016/j.jid.2018.12.018.
Zivanovic, V., S. Seifert, D. Drescher, P. Schrade, S. Werner, P. Guttmann, G. P. Szekeres, S. Bachmann, G. Schneider, C. Arenz, and J. Kneipp (2019). "Optical Nanosensing of Lipid Accumulation due to Enzyme Inhibition in Live Cells." ACS Nano 13(8): 9363-9375. ComorbSysMed doi.org/10.1021/acsnano.9b04001.



