MitoPD
Mitochondrial endophenotypes of PD
Parkinson disease (PD) is a progressive neurodegenerative disorder mainly affecting the motor system. Although early motor symptoms like tremor, rigidity, slowness of movement can be alleviated for several years, no disease-modifying treatment is available to date.
PD is an etiologically heterogeneous syndrome caused by a combination of genetic and environmental risk factors. Mutations that have conclusively been shown to cause familial forms of PD include mutations in genes like SNCA (α-synuclein), LRRK2 (leucine-rich repeat kinase 2), parkin, and PINK1 (figure 1).
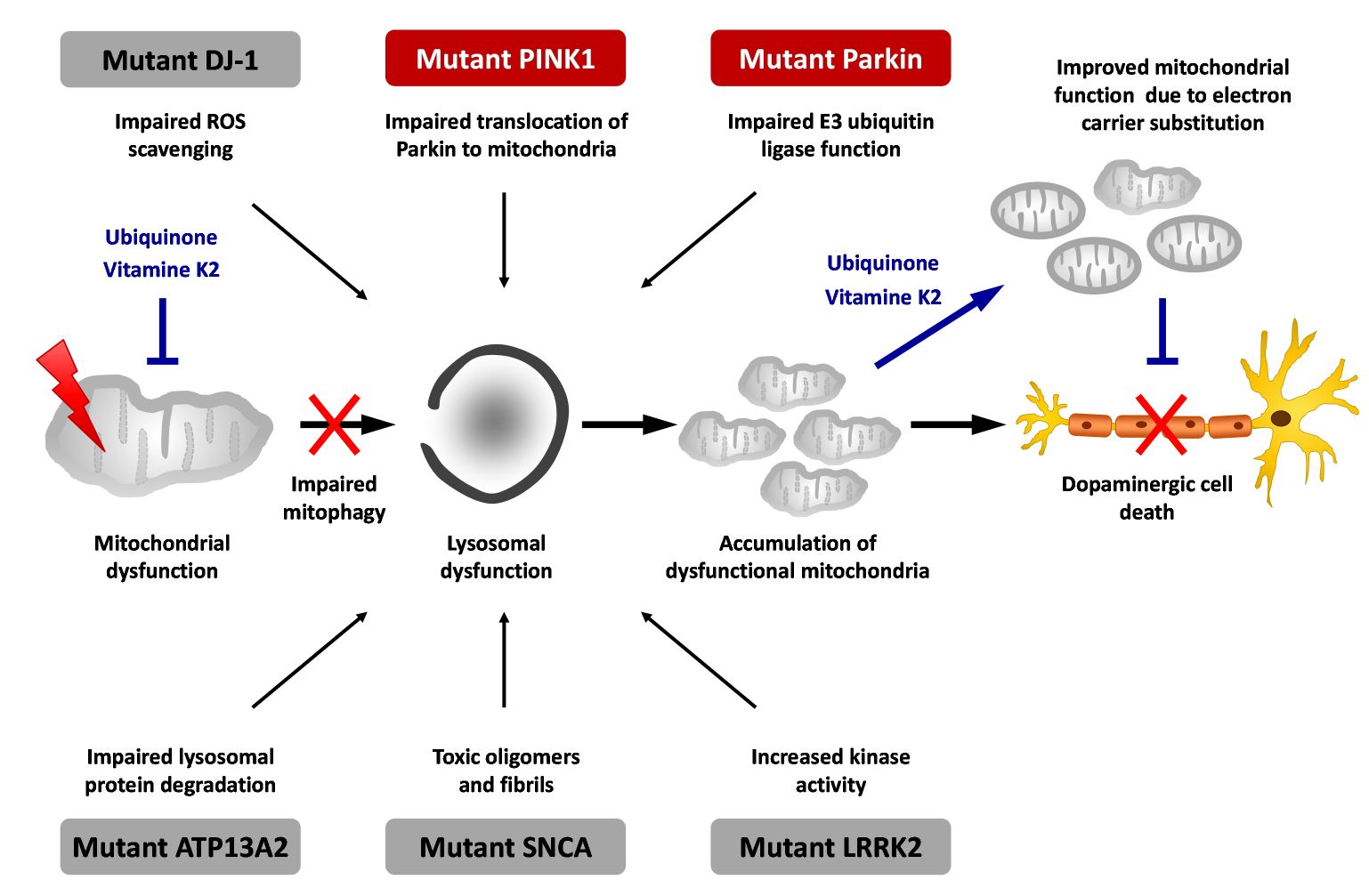

Genetic studies point to a limited number of molecular pathways likely to be involved in PD pathogenesis. Mitochondrial dysfunction and ensuing cellular energy failure and oxidative stress may be one of these crucial pathways, resulting in death of dopaminergic neurons. However, mitochondrial function is not the primary defect in all, but only in a subgroup of PD patients. We hypothesize that this etiologic heterogeneity of PD is one important cause for the failure to develop successful treatment so far.
Therefore, the goal of our project is to identify an etiologically homogeneous subgroup of PD, the “mitochondrial endophenotype”. The validity of this stratification process will be evaluated in cellular and animal models and by performing a proof-of-concept clinical trial, targeting the specific biochemical deficiency in this particular subgroup of patients.
In order to achieve this, a set of models will be generated from genomic, transcriptomic and (phospho)proteomic data from patients with known mitochondrial dysfunction (those with mutations in the genes encoding parkin and PINK1) and in corresponding animal and cellular models. These models will be used as prior information for model building in larger data sets from well characterized cohorts of patients with sporadic PD, allowing to sub-classify the cohorts according to predominant pathogenic pathways (figure 2).
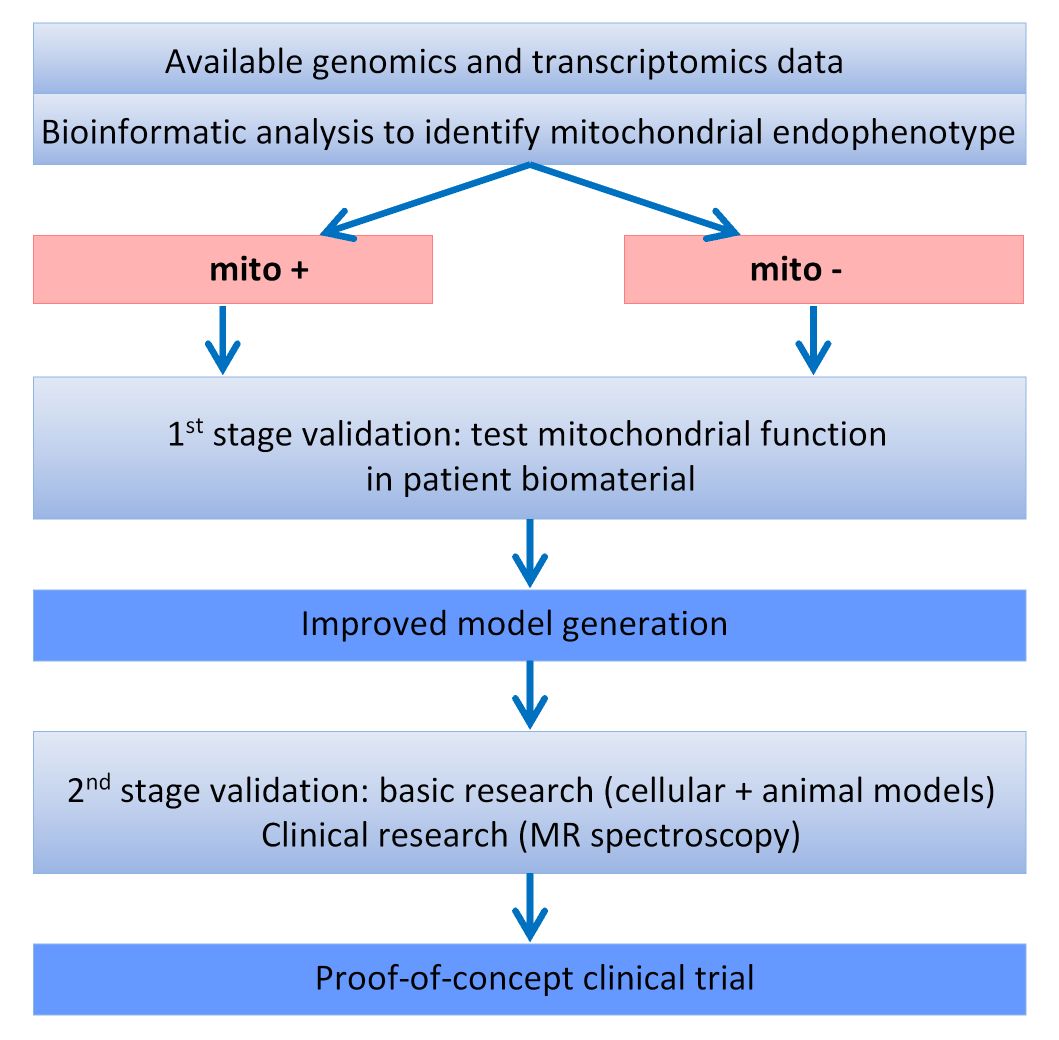
A first-stage validation will be performed by testing mitochondrial function in patient biomaterials. Results of this validation step will be used to improve model generation and validity will be confirmed in a second-stage validation in further genetically defined cohorts as well as in animal and cellular models of PD, including patient-derived induced pluripotent stem cells. Pathway-specific biomarkers will then be developed and used to define sub-cohorts that will be included in a proof-of-concept clinical trial using coenzyme Q10 and vitamine K2.
Subprojects in MitoPD:
SP 2 Data management and integration
SP 3 Computational modeling of mitochondrial dysfunction
SP 4 Validation in cellular models
SP 5 Mitochondrial endophenotypes of Parkinson's Disease
Keywords: Parkinson's disease, pathogenesis, mitochondria, treatment, systems biology



