SMOOSE
Systems-level analysis of modulators of oncogenic signaling
Oncology is witnessing a dramatic change of paradigm. The success of molecularly targeted cancer therapeutics in tumors with specific mutations has challenged the lack of progress through conventional cytotoxic chemotherapy. Thus, personalized healthcare has become a reality in the care of patients, exemplified by the growing number of tumors that are amenable to such approach (e.g. EGFR-mutant lung cancer, BRAF-mutant melanoma). We have established a framework for discovery and validation of therapeutically relevant genome alterations in cancer and we have implemented an outreach program to funnel such findings into routine clinical application. Unfortunately the success of personalized oncology is limited by acquired resistance and by primary resistance, elicited by additional genome alterations that modulate the responsiveness of the tumor to therapy. Here, we propose a multi-disciplinary approach – involving computer science and computational biology, molecular cancer biology and (epi)genomics, genetically manipulated mice, chemical and structural biology, molecular pathology and medical oncology. We thus wish to capture the genomic and signaling complexity of tumors in preclinical models and in patients, provide system-level models of evolutionary hierarchy in cancer genomes and the associated implications on pathway activation and drug response, develop preclinical models in cells and in mice to validate computational simulations and predictions, identify novel chemical routes for multi-pathway inhibition, and funnel our results into diagnostic approaches and clinical trials. Our efforts are based on several years of joint interdisciplinary work leading to successful translation of genomics discoveries into clinical trials. We now propose to expand our existing pipeline to globally capture the complex genotype-phenotype relationship that is reflective of the complex nature of cancer.
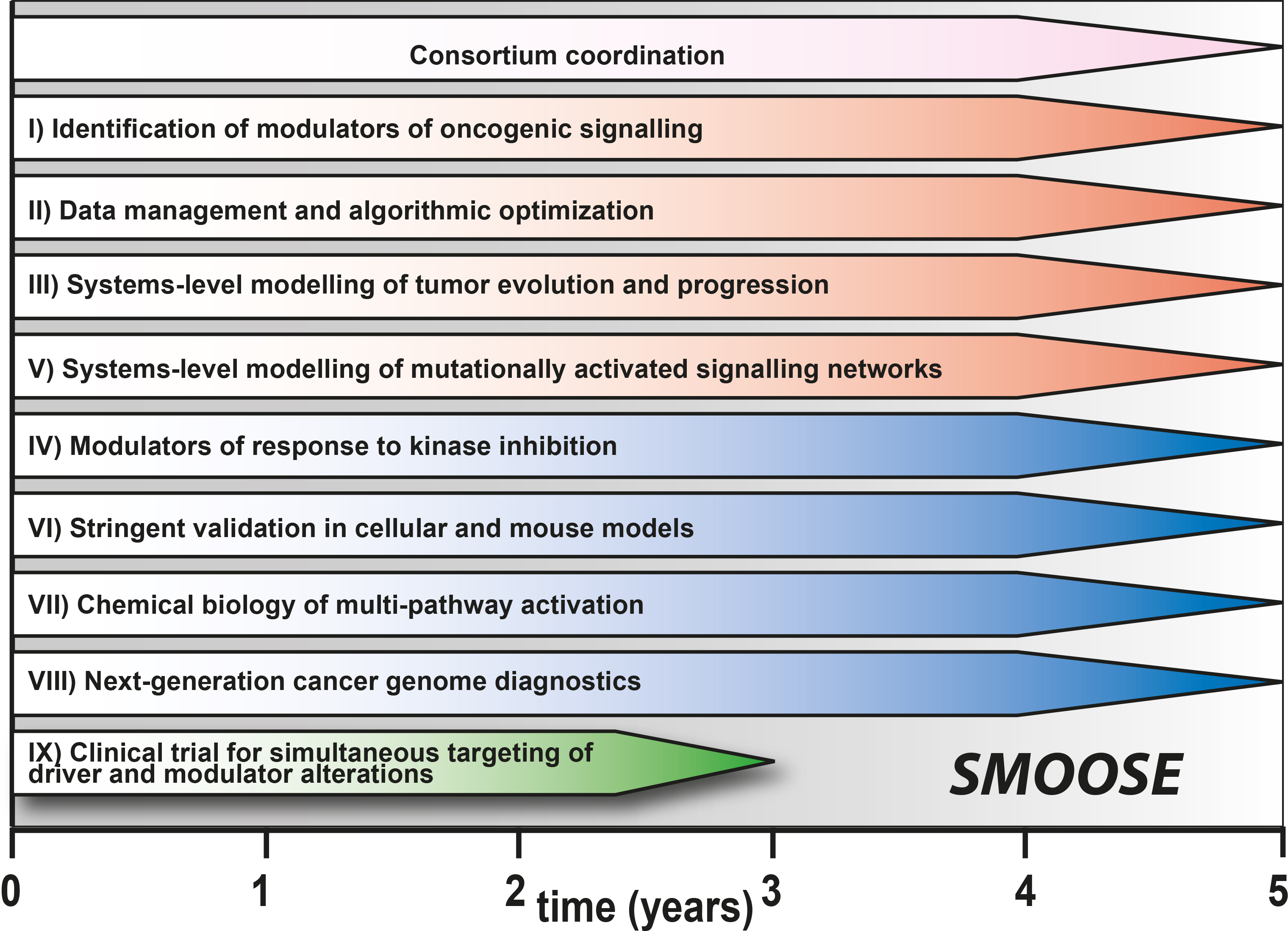

The goal of the overall consortium SMOOSE is to improve the quality of medicine for cancer patients by using system medicine for a comprehensive insight into the disease and its treatment. In contrast to isolated analysis of individual components of cancer, we aim to investigate the complex and multifactorial causes of lung cancer and neuroblastoma to identify modulators of oncogenic signaling.
Subprojects in SMOOSE:
SP 1 Genomic characterization and modeling of tumor progression
SP 2 Systems-level modeling of cancer genome evolution
SP 3 Systems-level modeling of mutationally activated signaling networks and response to therapy
SP 4 Data handling, optimization of analysis workflows and applications
SP 5 Identification, validation and exploitation of modulators
SP 6 In vivo characterization of oncogenically rewired signaling networks in lung cancer
SP 7 Modulation of oncogenic signaling through epigenetic writers of the histone code
SP 8 Chemical Biology of multi-pathway inhibition
Keywords: cancer research, genome, lung cancer, neuroblastoma



