SysMedOs
Integration of oxidative stress into systems medicine view for obesity and obesity related complications
Aims:
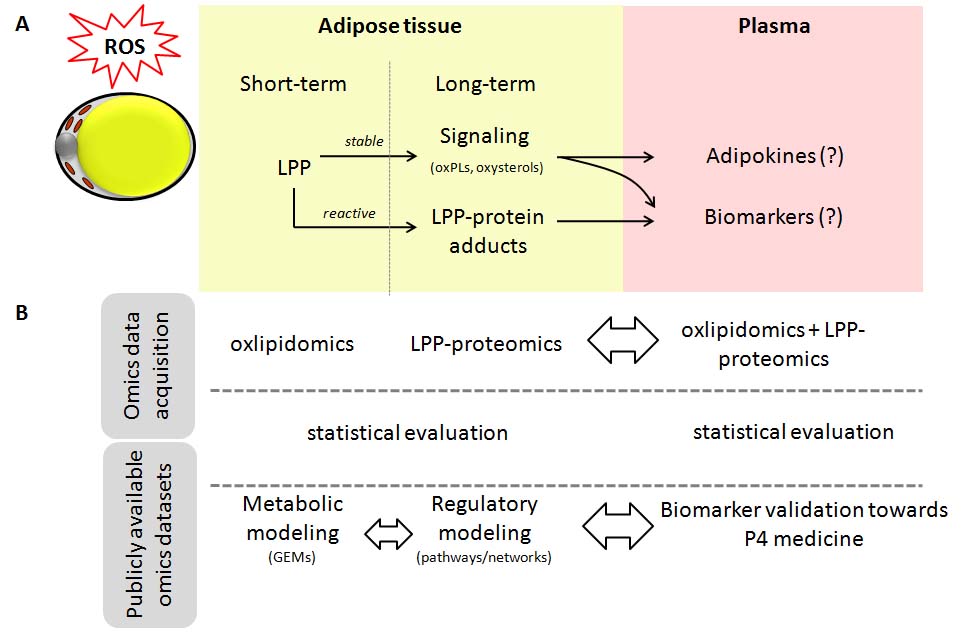
The project aims is to include knowledge on different oxidized versions of lipids and proteins into the integrated view on obesity and obesity related complications by systems medicine. The project will combine clinical parameters and omics data targeting oxidized lipids and lipid-modified proteins, as well as freely accessible multi-omics datasets in order to mathematically model the impact of oxidative stress on adipose tissue to distinguish metabolically healthy obese individuals from those with high metabolic risks. The proposed project goes far beyond the current strategies by integrating quantitative data obtained by oxlipidomics and proteomics of LPP-modified proteins (lipox-proteomics) from AT tissue and plasma samples in combination with publicly available omics datasets into mathematical models of adipose tissue and obesity.
Working plan:
The project will rely on combination of a wide array of approaches including omics data acquisition, clinical characterization, statistical evaluation, and mathematical modeling. Using previously published omics data and mathematical models of adipose tissue as a scaffold, newly generated data on oxidized lipids and proteins will be integrated via genome-scale metabolic modeling and regulatory protein networks analysis to obtain systems medicine view on metabolically healthy and diseased obesity phenotypes and identify potential biomarkers and therapeutic targets, which will be further validated in blood plasma samples of obese and lean individuals.
Impact:
The results will have high diagnostic, prognostic, and therapeutic potentials by providing significant impacts on our understanding of obesity outcomes and additionally on biomarker discovery and validation, design of new intervention therapies, and diagnostic/prognostic assays. These reliable biomarkers will push clinical developments by
- (i) accelerating disease progression monitoring and therapy evaluations,
- (ii) providing information on drug-target interactions, general changes in the pathophysiology of diseases and improvement of a clinical status, and
- (iii) reflecting possible side effects of therapeutic treatments.
- Moreover, the systematic view on obesity and obesity related complications assessable via enriched genome-scale metabolic models of AT (iAdipocyte1850) will guide the development of personalized, predictive, preventive, and participatory medicine (P4 Medicine).
Publications
Barayeu, U., M. Lange, L. Méndez, J. Arnhold, O. I. Shadyro, M. Fedorova, and J. Flemmig (2019). "Cytochrome c auto-catalyzed carbonylation in the presence of hydrogen peroxide and cardiolipins." The Journal of Biological Chemistry 294(6): 1816-1830. SysMedOs doi.org/10.1074/jbc.RA118.004110.
Bhandari, D. R., G. Coliva, M. Fedorova, and B. Spengler (2020). "Single Cell Analysis by High-Resolution Atmospheric-Pressure MALDI MS Imaging." Methods Mol Biol 2064: 103-111. SysMedOS doi.org/10.1007/978-1-4939-9831-9_8.
Brakel, A., D. Volke, C. N. Kraus, L. Otvos, and R. Hoffmann (2019). "Quantitation of a Novel Engineered Anti-infective Host Defense Peptide, ARV-1502: Pharmacokinetic Study of Different Doses in Rats and Dogs." Front Chem 7: 753. SysMedOs doi.org/10.3389/fchem.2019.00753.
Campos-Pinto, I., L. Mendez, J. Schouten, J. Wilkins, M. Fedorova, A. Pitt, P. Davis, and C. Spickett (2019). "Epitope mapping and characterization of 4-hydroxy-2-nonenal modified-human serum albumin using two different polyclonal antibodies." Free Radic Biol Med. SysMedOs doi.org/10.1016/j.freeradbiomed.2019.05.008.
Coliva, G., S. Duarte, D. Perez-Sala, and M. Fedorova (2019). "Impact of inhibition of the autophagy-lysosomal pathway on biomolecules carbonylation and proteome regulation in rat cardiac cells." Redox Biol: 101123. SysMedOS doi.org/10.1016/j.redox.2019.101123.
Colombo, S., A. Criscuolo, M. Zeller, M. Fedorova, M. R. Domingues, and P. Domingues (2019). "Analysis of oxidised and glycated aminophospholipids: Complete structural characterisation by C30 liquid chromatography-high resolution tandem mass spectrometry." Free Radic Biol Med 144: 144-155. SysMedOs doi.org/10.1016/j.freeradbiomed.2019.05.025.
Criscuolo, A., P. Nepachalovich, D. F. Garcia-del Rio, M. Lange, Z. Ni, M. Baroni, G. Cruciani, L. Goracci, M. Blüher, and M. Fedorova (2022). "Analytical and computational workflow for in-depth analysis of oxidized complex lipids in blood plasma." Nat Commun 13(6547): 1–13. SysMedOS doi.org/10.1038/s41467-022-33225-9.
Criscuolo, A., M. Zeller, K. Cook, G. Angelidou, and M. Fedorova (2019). "Rational selection of reverse phase columns for high throughput LC-MS lipidomics." Chem Phys Lipids 221: 120-127. SysMedOs doi.org/10.1016/j.chemphyslip.2019.03.006.
Criscuolo, A., M. Zeller, and M. Fedorova (2020). "Evaluation of Lipid In-Source Fragmentation on Different Orbitrap-based Mass Spectrometers." J. Am. Soc. Mass Spectrom. SysMedOs doi.org/10.1021/jasms.9b00061.
Gaud, C., B. C. Sousa, A. Nguyen, M. Fedorova, Z. Ni, V. B. O’Donnell, M. J. O. Wakelam, S. Andrews, and A. F. Lopez-Clavijo (2021). "BioPAN: a web-based tool to explore mammalian lipidome metabolic pathways on LIPID MAPS." F1000Research 10: 4. SysMedOS f1000research.com/articles/10-4/v2.
Koelmel, J. P., J. J. Aristizabal-Henao, Z. Ni, M. Fedorova, S. Kato, Y. Otoki, K. Nakagawa, E. Z. Lin, K. J. Godri Pollitt, V. Vasiliou, J. D. Guingab, T. J. Garrett, T. L. Williams, J. A. Bowden, and M. Penumetcha (2021). "A Novel Technique for Redox Lipidomics Using Mass Spectrometry: Application on Vegetable Oils Used to Fry Potatoes." J Am Soc Mass Spectrom. SysMedOS www.ncbi.nlm.nih.gov/pubmed/34096708.
Kofeler, H. C., T. O. Eichmann, R. Ahrends, J. A. Bowden, N. Danne-Rasche, E. A. Dennis, M. Fedorova, W. J. Griffiths, X. Han, J. Hartler, M. Holcapek, R. Jirasko, J. P. Koelmel, C. S. Ejsing, G. Liebisch, Z. Ni, V. B. O'Donnell, O. Quehenberger, D. Schwudke, A. Shevchenko, M. J. O. Wakelam, M. R. Wenk, D. Wolrab, and K. Ekroos (2021). "Quality control requirements for the correct annotation of lipidomics data." Nat Commun 12(1): 4771. SysMedOS www.ncbi.nlm.nih.gov/pubmed/34362906.
Kreiter, J., A. Rupprecht, L. Zimmermann, M. Moschinger, T. I. Rokitskaya, Y. N. Antonenko, L. Gille, M. Fedorova, and E. E. Pohl (2019). "Molecular Mechanisms Responsible for Pharmacological Effects of Genipin on Mitochondrial Proteins." Biophys J 117(10): 1845-1857. SysMedOS doi.org/10.1016/j.bpj.2019.10.021.
Kyle, J. E., L. Aimo, A. J. Bridge, G. Clair, M. Fedorova, J. B. Helms, M. R. Molenaar, Z. Ni, M. Oresic, D. Slenter, E. Willighagen, and B. M. Webb-Robertson (2021). "Interpreting the lipidome: bioinformatic approaches to embrace the complexity." Metabolomics 17(6): 55. SysMedOS www.ncbi.nlm.nih.gov/pubmed/34091802.
Lange, M., and M. Fedorova (2020). "Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma." Anal Bioanal.Chem: 1–12. SysMedOs doi.org/10.1007/s00216-020-02576-x.
Lange, M., Z. Ni, A. Criscuolo, and M. Fedorova (2018). "Liquid Chromatography Techniques in Lipidomics Research." Chromatographia. SysMedOs doi.org/10.1007/s10337-018-3656-4.
Mishima, E., J. Ito, Z. Wu, T. Nakamura, A. Wahida, S. Doll, W. Tonnus, P. Nepachalovich, E. Eggenhofer, M. Aldrovandi, B. Henkelmann, K.-i. Yamada, J. Wanninger, O. Zilka, E. Sato, R. Feederle, D. Hass, A. Maida, A. S. D. Mourão, A. Linkermann, E. K. Geissler, K. Nakagawa, T. Abe, M. Fedorova, B. Proneth, D. A. Pratt, and M. Conrad (2022). "A non-canonical vitamin K cycle is a potent ferroptosis suppressor." Nature 608: 778–783. SysMedOS doi.org/10.1038/s41586-022-05022-3.
Narzt, M. S., I. M. Nagelreiter, O. Oskolkova, V. N. Bochkov, J. Latreille, M. Fedorova, Z. Ni, F. J. Sialana, G. Lubec, M. Filzwieser, M. Laggner, M. Bilban, M. Mildner, E. Tschachler, J. Grillari, and F. Gruber (2019). "A novel role for NUPR1 in the keratinocyte stress response to UV oxidized phospholipids." Redox Biol 20: 467-482. SysMedOS doi.org/10.1016/j.redox.2018.11.006.
Ni, Z., G. Angelidou, R. Hoffmann, and M. Fedorova (2017). "LPPtiger software for lipidome-specific prediction and identification of oxidized phospholipids from LC-MS datasets." Scientific Reports 7(1): 15138. SysMedOs doi.org/10.1038/s41598-017-15363-z.
Ni, Z., G. Angelidou, M. Lange, R. Hoffmann, and M. Fedorova (2017). "LipidHunter Identifies Phospholipids by High-Throughput Processing of LC-MS and Shotgun Lipidomics Datasets." Analytical Chemistry 89(17): 8800-8807. SysMedOS doi.org/10.1021/acs.analchem.7b01126.
Ni, Z., L. Goracci, G. Cruciani, and M. Fedorova (2019). "Computational solutions in redox lipidomics - Current strategies and future perspectives." Free Radic Biol Med. SysMedOS doi.org/10.1016/j.freeradbiomed.2019.04.027.
Ni, Z., B. C. Sousa, S. Colombo, C. B. Afonso, T. Melo, A. R. Pitt, C. M. Spickett, P. Domingues, M. R. Domingues, M. Fedorova, and A. Criscuolo (2019). "Evaluation of air oxidized PAPC: A multi laboratory study by LC-MS/MS." Free Radic Biol Med. SysMedOs doi.org/10.1016/j.freeradbiomed.2019.06.013.
Sochorová, M., K. Vávrová, M. Fedorova, Z. Ni, D. Slenter, M. Kutmon, E. L. Willighagen, S. Letsiou, D. Töröcsik, M. Marchetti-Deschmann, S. Zoratto, C. Kremslehner, and F. Gruber (2022). "Research Techniques Made Simple: Lipidomic Analysis in Skin Research." J Invest Dermatol 142(1): 4–11.e11. SysMedOS doi.org/10.1016/j.jid.2021.09.017.



