CLIOMMICS
Clinically-applicable, omics-based assessment of survival, side effects, and targets in multiple myeloma
Multiple Myeloma (MM) for the most part is an incurable malignant plasma cell disease with an incidence rate of 4-8 per 100,000 inhabitants. Clonal plasma cells accumulate in the bone marrow causing clinical signs and symptoms based on the displacement of normal hematopoiesis and the formation of osteolytic bone lesions or osteopenia. Myeloma cells harbor a high median number of chromosomal aberrations, and multiple changes in gene expression compared to normal plasma cells. The prognosis of MM patients has significantly been improved during the last decade due to the introduction of novel agents into treatment and is now approaching 10 years. While there is no cure for Multiple Myeloma in most patients, the cancer can be managed successfully in many patients for years and yet an increasingly number of complete remissions of the disease at all stages of treatment were reported. Concomitantly Multiple Myeloma and its treatment often cause therapy-limiting side effects like peripheral polyneuropathy and hematological toxicity.
The challenges in understanding and treatment of Multiple Myeloma are complex and can only be addressed in a system-oriented approach. Within the consortium CLIOMMICS between University Hospital Heidelberg and German Cancer Research Center Heidelberg (DKFZ) data already available, or still to be gathered, for the prediction of progression free survival (transcriptomics, B3), prediction of side effects (GWAS, B2) and assessment of targets individually present only in a sub-fraction of patients (B3) are integrated and subsequently affiliated with conventional clinical factors and imaging data (B4). The essential and extensive bioinformatics background for data integration and implementation of reporting is done in B1.
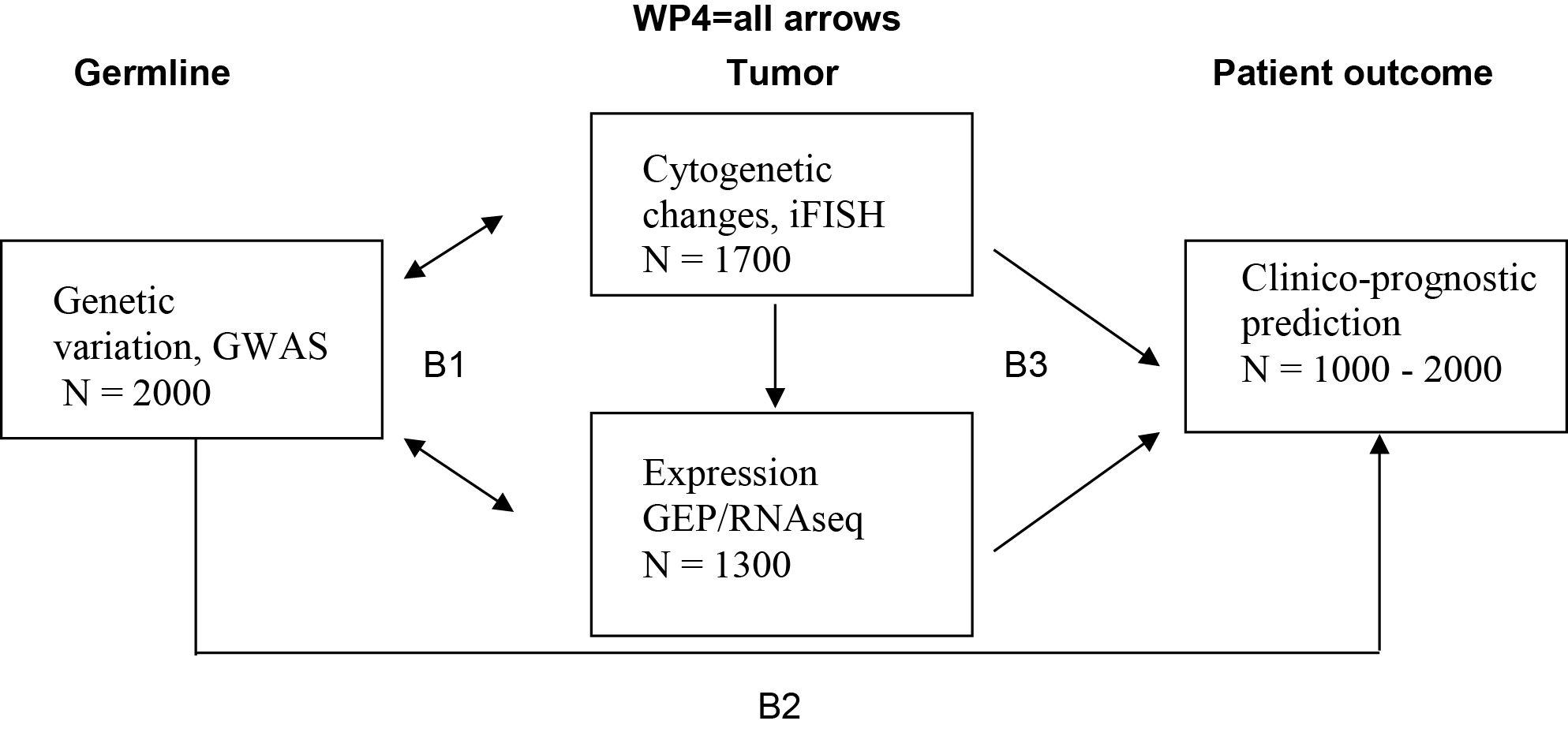
The interrelation between different “omics”, molecular and clinical prognostic factors will help to predict survival, response to treatment or therapy-limiting side effects and will lead to a complete novel level of understanding of the disease. Reporting of comprehensive data within a timespan and in a validated way will for the first time lead to an individual CLIOMMICS patient report allowing clinical decisions to be made. The overall goal is to personalize myeloma treatment, thereby improving the long term survival and quality of life for myeloma patients.
Subprojects in CLIOMMICS:
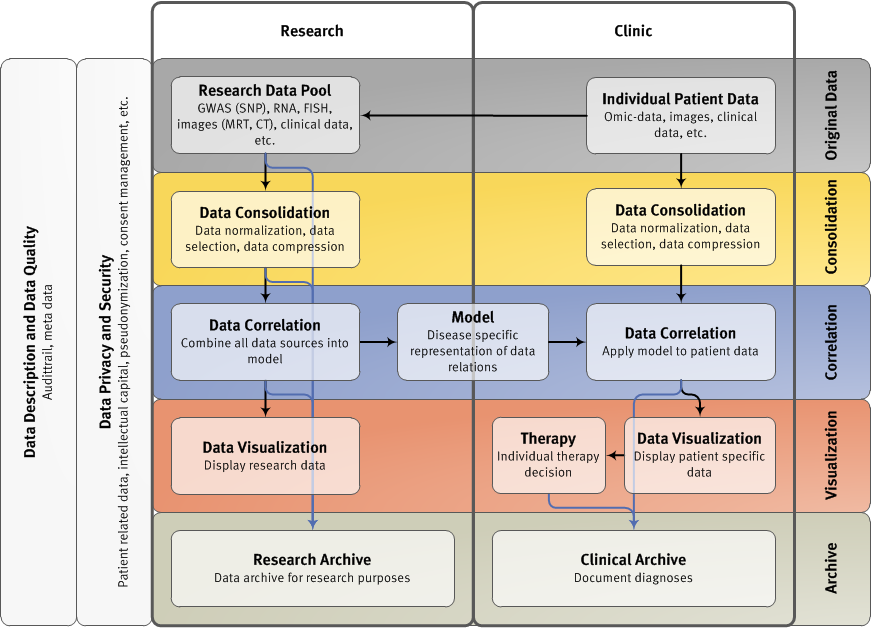
SP 1 IT-Architecture and Multi-level Data Management for Systems Medicine for Multiple Myeloma
SP 3 Transcriptomics by RNA-sequencing: Performing and reporting in clinical routine
Keywords: Multiple Myeloma, response to treatment, side effects, prognosis, gene expression, HM-metascore, integrative analysis, prediction of course of disease, patient report, multi level data management, IT architecture, systems medicine



