Sys_CARE
Systems medicine investigation of alternative splicing in Cardiac and Renal Diseases
Alternative splicing (AS) is a biological mechanism by which different transcripts (RNA copies of a DNA segment) and ultimately different proteins can be formed from the same DNA sequence (gene). AS is known to be causally involved in several diseases. Aberrations of AS can influence the cellular machinery on multiple molecular scales, albeit the impact of each individual event is likely to be subtle. A systematic analysis of the association between alterations in AS and particular diseases is still lacking.
To tackle this, Sys_CARE brings together expertise from biomedical research and clinical practise (University Medicine Greifswald) as well as computational biology (Universität Hamburg and Helmholtz Institute for Pharmaceutical Research Saarland) to establish the first systems medicine view on AS.
Sys_CARE will analyze a large set of multi-Omics data in the context of molecular interaction networks. As a first step, transcripts will be identified that are significantly more abundant in patients affected by two major human diseases, dilated cardiomyopathy (DCM) and hypertensive nephrosclerosis (HN). Alternatively spliced isoforms will be characterized with a focus on their potential to form protein complexes. Based on this, we will predict changes in protein-protein interaction networks affecting disease development. De novo network enrichment methods will allow us to integrate gene expression data with these networks to study how AS affects cellular mechanisms and regulatory programs.
In parallel, we will investigate AS events in time-course studies using mouse and zebrafish models of DCM and HN, respectively. This will allow us to directly compare insights across organisms and to validate our findings with temporal resolution of disease development, in order to test if and how AS is causally involved.
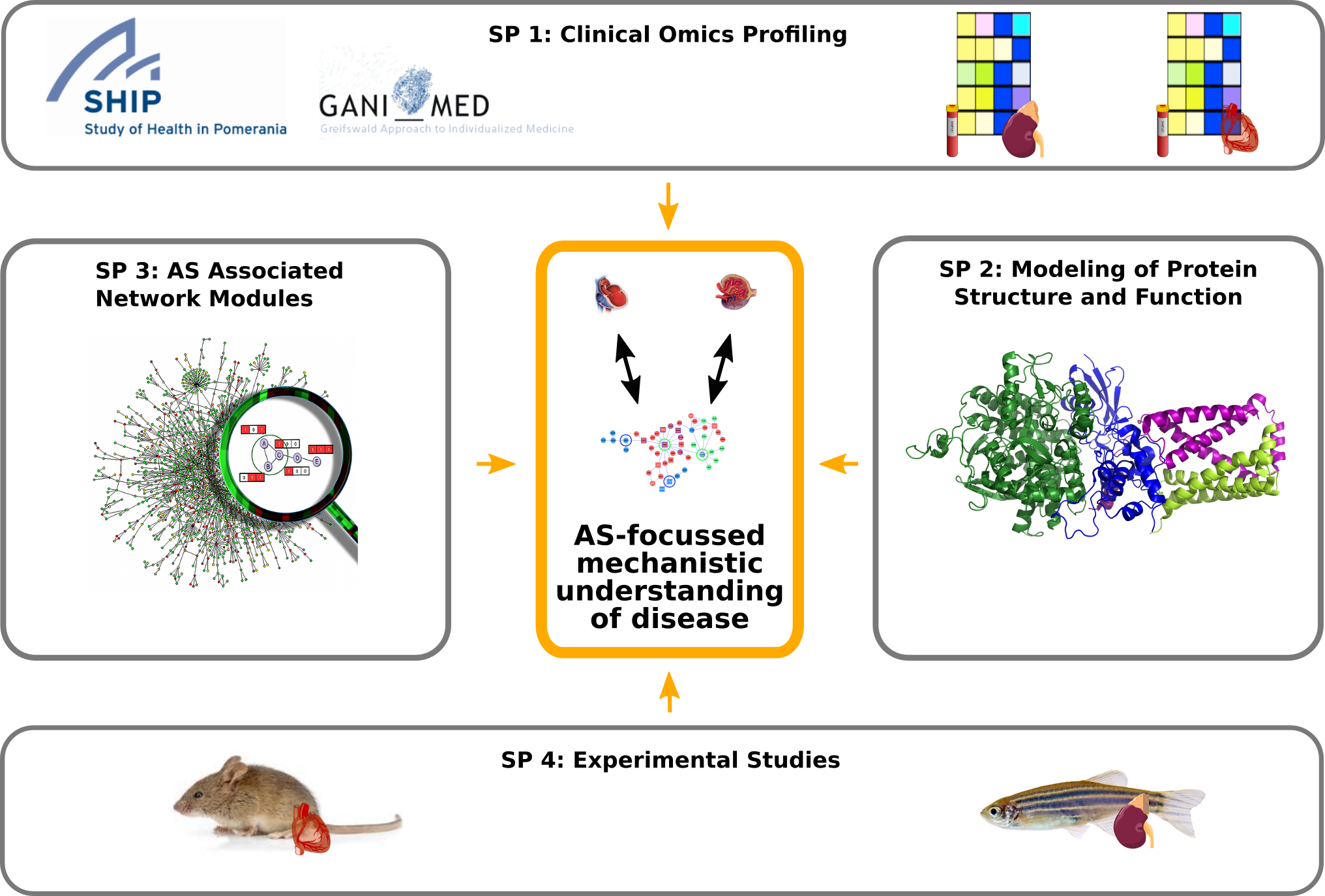

Project organization in Sys_CARE: interactions of four different subprojects focused on the identification of alternative splicing associated network modules.
Finally, the results obtained in silico will be verified in vitro and in vivo. Experimental results will be used to iteratively refine our network analysis methodology. We expect that the insights we gain in Sys_CARE regarding the role of AS in DCM and HN will have translational potential for the advancement of precision medicine, as AS can be influenced therapeutically, e.g. via antisense oligonucleotides.
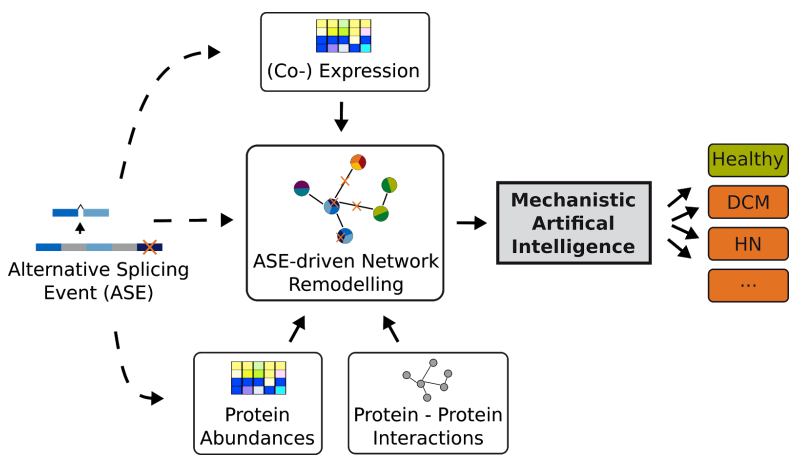
The Sys_CARE project aims at understanding disease-associated alternative splicing events on a mechanistic level. DCM and HN cohorts are compared to a population-based cohort (SHIP) to identify common as well as unique alternative splicing events. Additionally, animal models will be investigated temporally to gain insights into disease progression.



