SP 2
Mathematical large-scale modelling of signaling pathways in pancreatic neuroendocrine tumors (pNET)
Patient-individual biological changes (mutation, expression, activation) will be mapped in a mathematical MAPK / PI3K / mTOR model. This model will then be able to make accurate predictions if available, or to be used in the future therapeutic interventions in these pathways are successful, or whether "escape mechanisms" in the sense of resistance to therapy should be ecpected. For this different genetic alterations and differential expression (protein/gene) are correlated a) with dynamic MAPK / mTOR signatures, and b) with differential treatment responses.
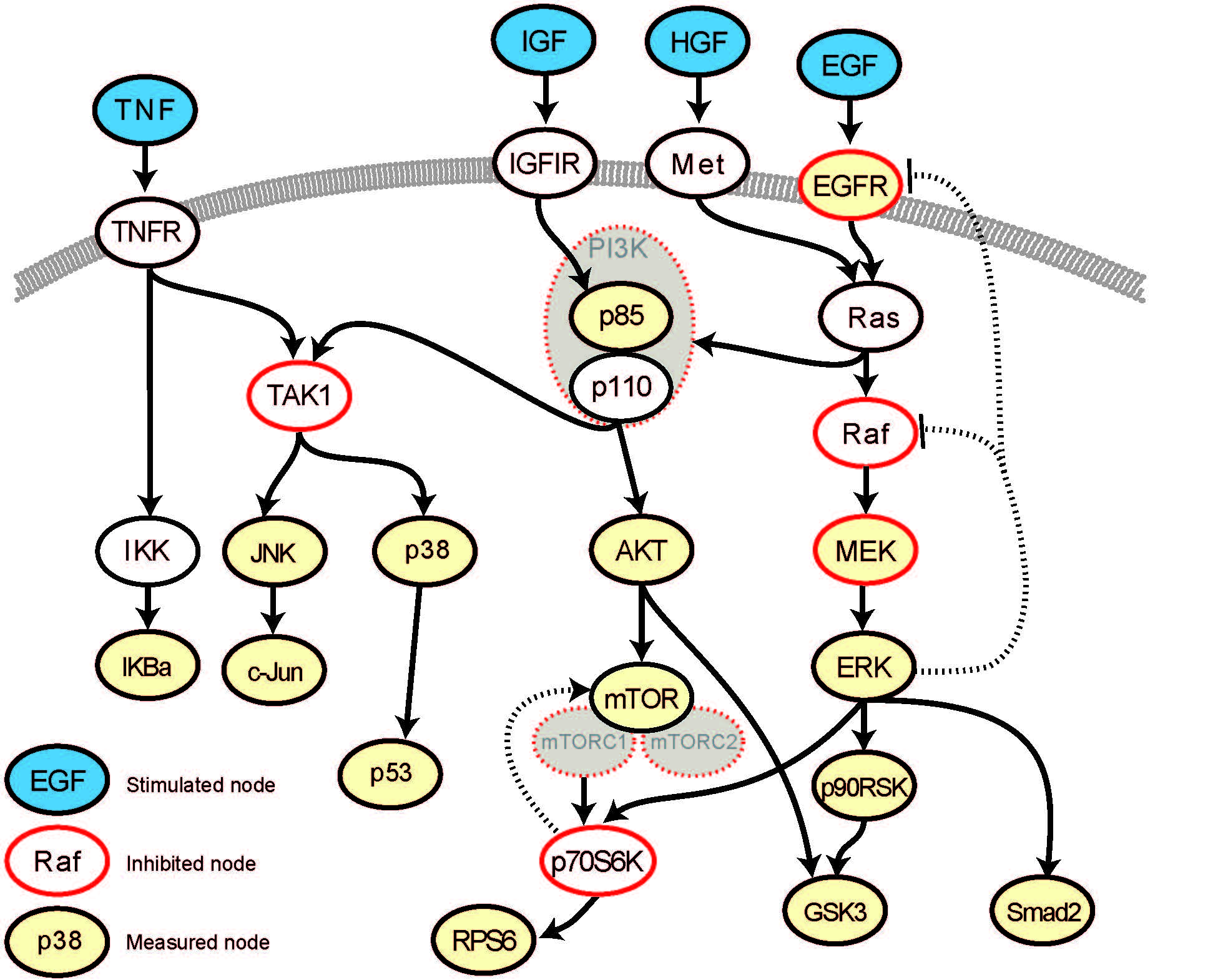
The main task of this subproject is the modelling and simulation patient-specific models of MAPK/PI3K/mTOR pathways. Technically, the model will be developed by (i) connecting the existing models of MAPK/PI3K signaling with a mTOR-pathway model and adaptation to neuroendocrine tumor cells, (ii) adapting the model parameters using time-series and perturbation data sets to generate models that can simulate the pathways/signal transduction under specific mutation pattern, and (iii) further expanding the models towards important nodes identified during the project.
The models are first generated with an already in the AG Blüthgen established Method of Modular-response analysis, but then extended by so-called regression models to parameterize larger network models. Aspects, in which the time-dynamics of the signaling networks are considered to be important, are represented by differential-equation models.
In addition, the effects of signaling networks in the phenotypes of cells (proliferation, migration, cell death) are modeled using regression models or other appropriate statistical models to predict the clinically relevant processes.
In addition, Nils Blüthgen covers the statistical data analysis and develops a model-assisted experimental design to optimize the experiments in the other subprojects (Sers AG and AG Detjen) thereby optimizing the model-experiment-cycles. The model will then be able to generate model predictions that can then be verified in various experimental models.
Keywords: mathematical MAPK / PI3K / mTOR model, Modular-response analysis, differential-equation models



