SP 4
Cell physiology modelling and proteomics
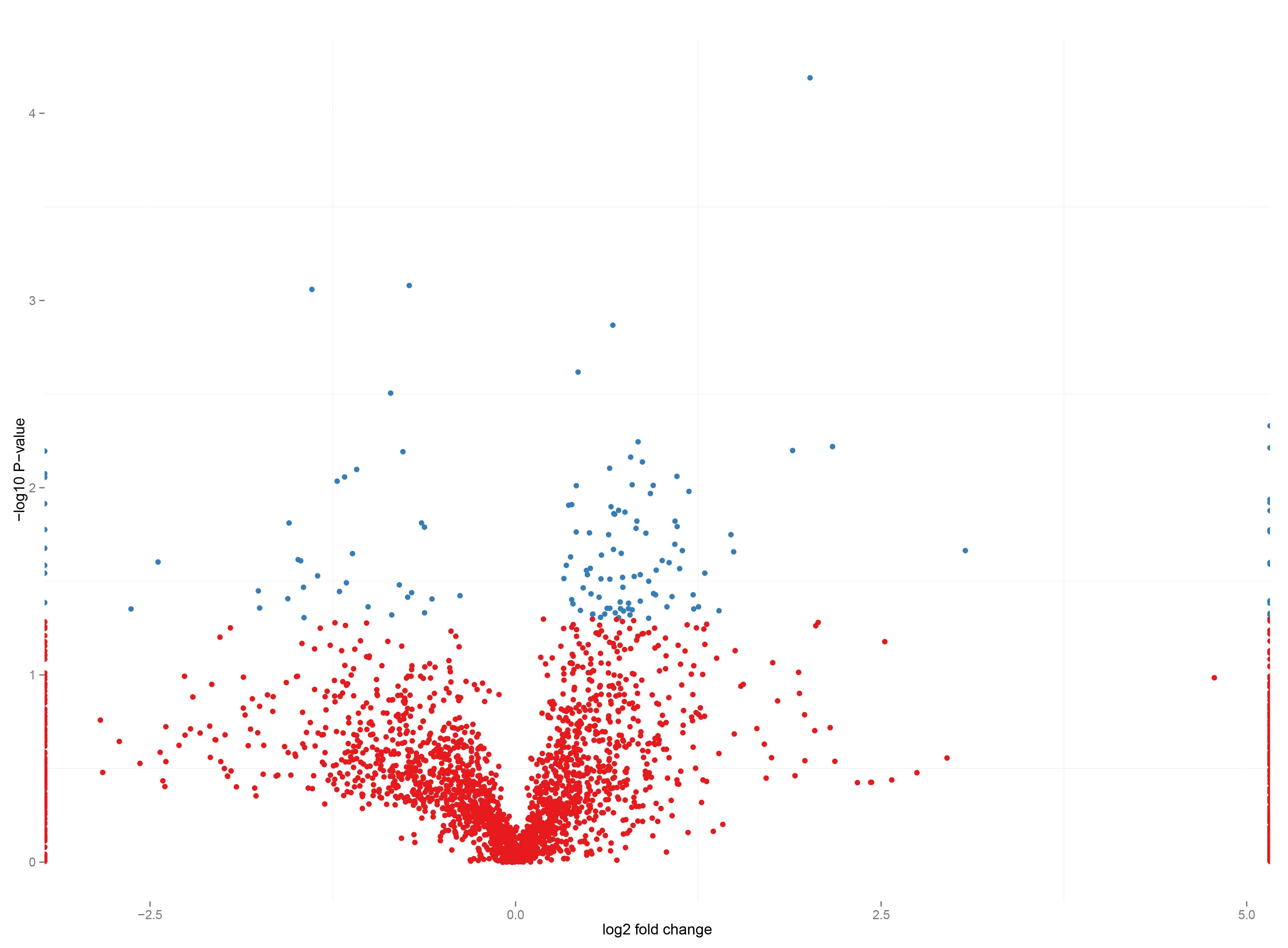
Proteomic analysis of patient samples determines the concentration of thousands of proreins individually for each patient. We thus relate cellular parameters to the individual disease profile. Mathematical modelling relates protein concentrations to function by simulating contraction with the individual parameter sets. As the case may be, it indicates gaps in the quantification of patient samples and/or in our understanding of disease mechanisms.

The work schedule comprises:
- development of a detailed mathematical model of excitation contraction coupling
- proteomic analysis of patient smaples
- simulation of patientspecific cell function
- comparison/verification with patient specific function data from imaging