SP3
Computational modeling of mitochondrial dysfunction
This project will apply computational analyses to large-scale biomolecular and genomic datasets from Parkinson’s disease (PD) patients and control individuals in order to identify and characterize sub-groups of patients with disease-associated alterations in the mitochondrium (the cell organelle responsible for energy production, thought to become dysfunctional in a large fraction of sporadic PD patients). The bioinformatic analyses will enable a more detailed characterization of the PD subtypes associated with mitochondrial dysfunction and the proposal of potential diagnostic approaches to discriminate these sub-groups against other PD subtypes and unaffected controls. This work could also provide a basis for the subsequent preclinical development of subtype-specific therapeutic strategies to slow down the progression of the disease or alleviate its symptoms.
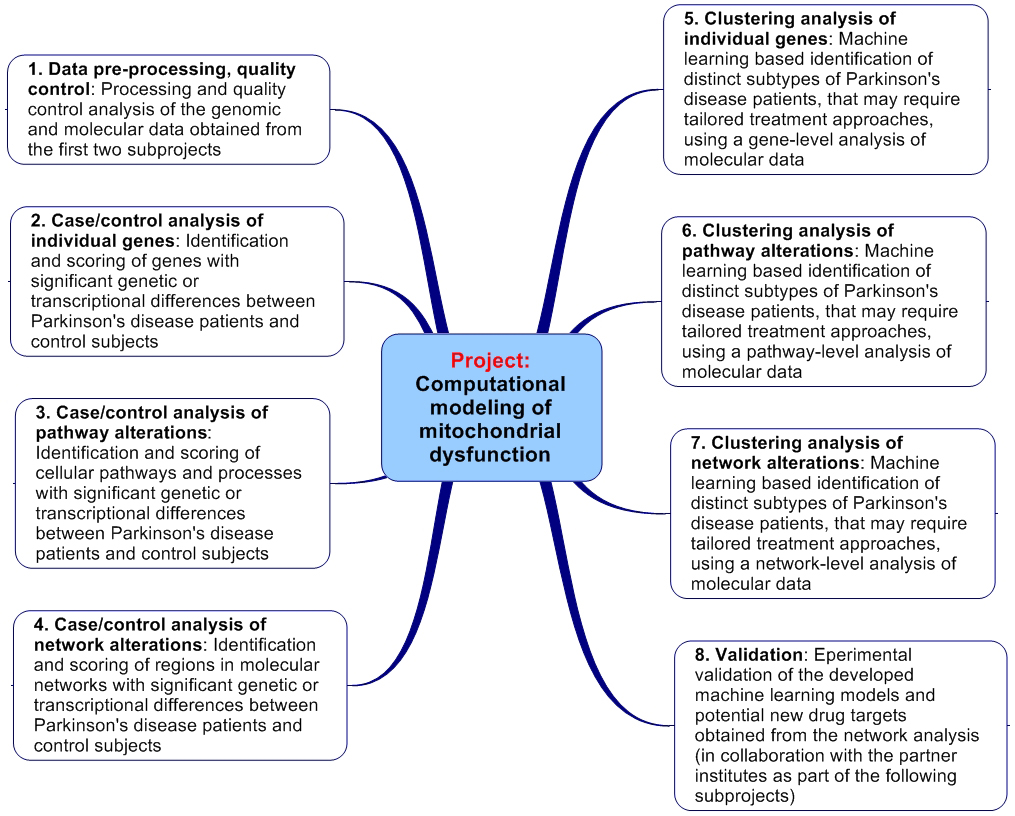
First, mitochondrial biomolecular and genetic differences between PD patients and controls, and between putative sub-groups of PD patients, will be identified and scored according to their statistical significance using bioinformatic analyses operating at the level of single genes/proteins, cellular signaling and metabolic pathways, and cellular networks of interacting biomolecules. Optimization techniques from the field of operations research will be used to identify the regions in molecular networks which display the strongest disease-associated alterations.
Next, we will compare characteristic deregulations in the identified sample groups for sporadic PD patients with those observed in patients with a known genetic cause of the disease associated with mitochondrial dysfunction (i.e. patients with a mutation in the genes PINK1 and PARK2). The main goal is to provide biologically interpretable, predictive machine learning models, which use information from molecular activity alterations in cellular pathways and networks to discriminate biological specimen from distinct PD-subtypes and unaffected controls. Moreover, the network alteration analyses will provide information relevant for prioritizing existing therapeutic strategies and proposing alternative drug targets. Finally, as part of complementary projects by the collaborating partners, the diagnostic models and proposed targets will be evaluated experimentally.



