SP 1
Validating the predictive SYSACT model under organotypic 3D conditions using TRAIL/IZI1551-derivatives and trametinib/dabrafenib
The data- and pathway knowledge-driven mathematical model (SYSACT) can accurately predict TRAIL and dacarbacine responsiveness of melanoma cell lines. Merging the quantitative expression pattern of 17 apoptosis regulators with topological information on apoptotic signal transduction allowed to generate a knowledge base from which responsiveness can be predicted.
We aim to translate predictions to more complex conditions, by integrating mature melanoma spheroids, which represent an intra-tumoral differentiation similar to avascularized cutaneous metastasis, into a 3D organotypic skin model. Spheroids will be generated from cell lines that within SYSACT have proven to be sensitive, low responding or resistant to TRAIL. Spheroids will be analysed in vitro as well as integrated into the 3D organotypic host tissue. Susceptibility to TRAIL will be determined immunhistochemically using paraffin sections and the expression of the decisive proteins, identified by SYSACT determined by standardized luminescence immunodetection (see WP3). Results will be integrated back to tune the SYSACT model, allowing to identify the most sensitive proteins in an organotypic environment that may serve as predictive marker.
Thereby, conventional TRAIL will be replaced by a new class of TRAIL-receptor agonists that overcome the intrinsic limitation observed for therapeutic antibodies (IZI1551, hexavalent Fc-scTRAIL fusion proteins; see WP4). This will allow investigating the predictive power of the model in 3D and the therapeutic improvement of Fc-scTRAIL in killing melanoma. Vice versa, quantitative expression pattern of the 17 apoptosis regulating proteins identified by SYSACT will be investigated in melanoma cells tested regarding their TRAIL sensitivity under 3D conditions.
In parallel identical experiments will be performed using specific BRAF (dabrafenib)/MEK (trametinib) inhibitors as single agents and combined with Fc-scTRAIL. In collaboration with WP2 BRAFV600E and BRAFwt melanoma cell lines with a known Ras mutation status will be evaluated in 2D and 3D. Special emphasis will be given to the quantification of proteins identified by Boolean modelling as sensitive parameter in the TRAIL and MEK signaling network (see WP2).
We aim to reduce the number of proteins needed to predict susceptibility towards combination treatments to a minimum, to serve as biomarkers for melanoma treatment for personalized medicine.
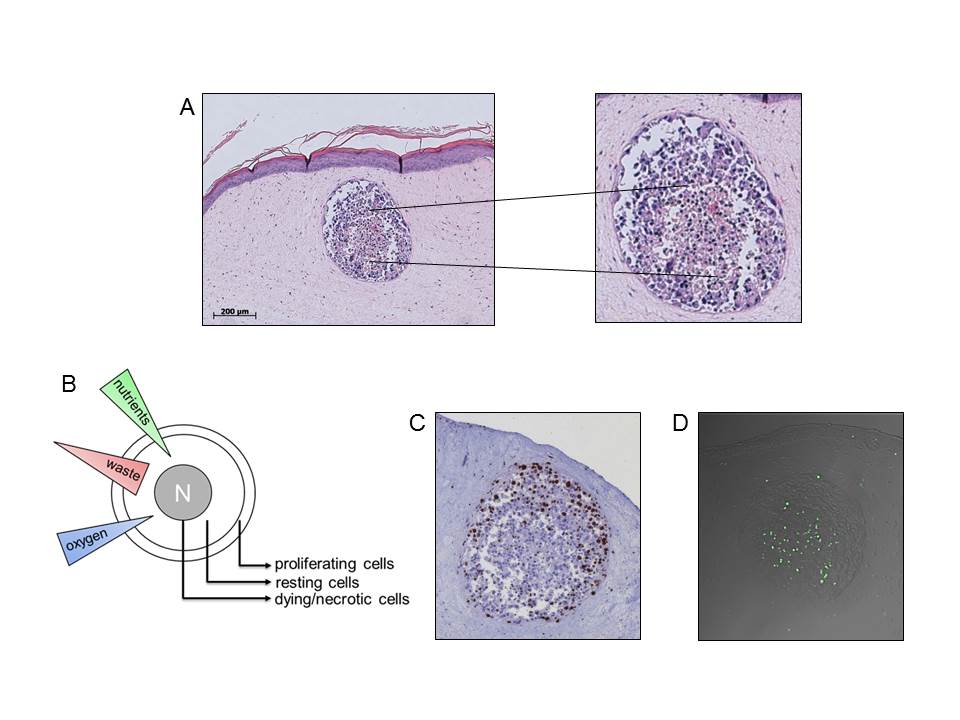
A Spheroids consist of at least two sub-populations, peripheral differentiated cells and pyknotic cells in the center. B Sub-populations of avasularized metastasis develop due to unequal distribution of nutrients, waste and oxygen. C Staining of cell cycle active Ki-67 protein: peripheral cells keep the proliferation capacity. D Staining of DNA strand break via TUNEL method: the spheroid core contains apoptoticx cells. Vörsmann et al., Cell Death Dis, 2013
Keywords: melanoma, apoptosis, signal transduction, systems biology



