SP 3
Functional Genomics in Human Heart Failure
This project within the research consortium aims to elucidate the interplay between heart failure and gene regulatory mechanisms by complex networks (transcriptome, miRnome, and genome data/profiles) using a large biobank of diseased human heart samples. We will define genetic variants, regulating microRNAs as well as key transcriptome data to pin down associated with poor clinical outcome. We will also use patient-specific models (mouse and iPS cell lines) to confirm networks that are involved in the pathogenesis of disease.
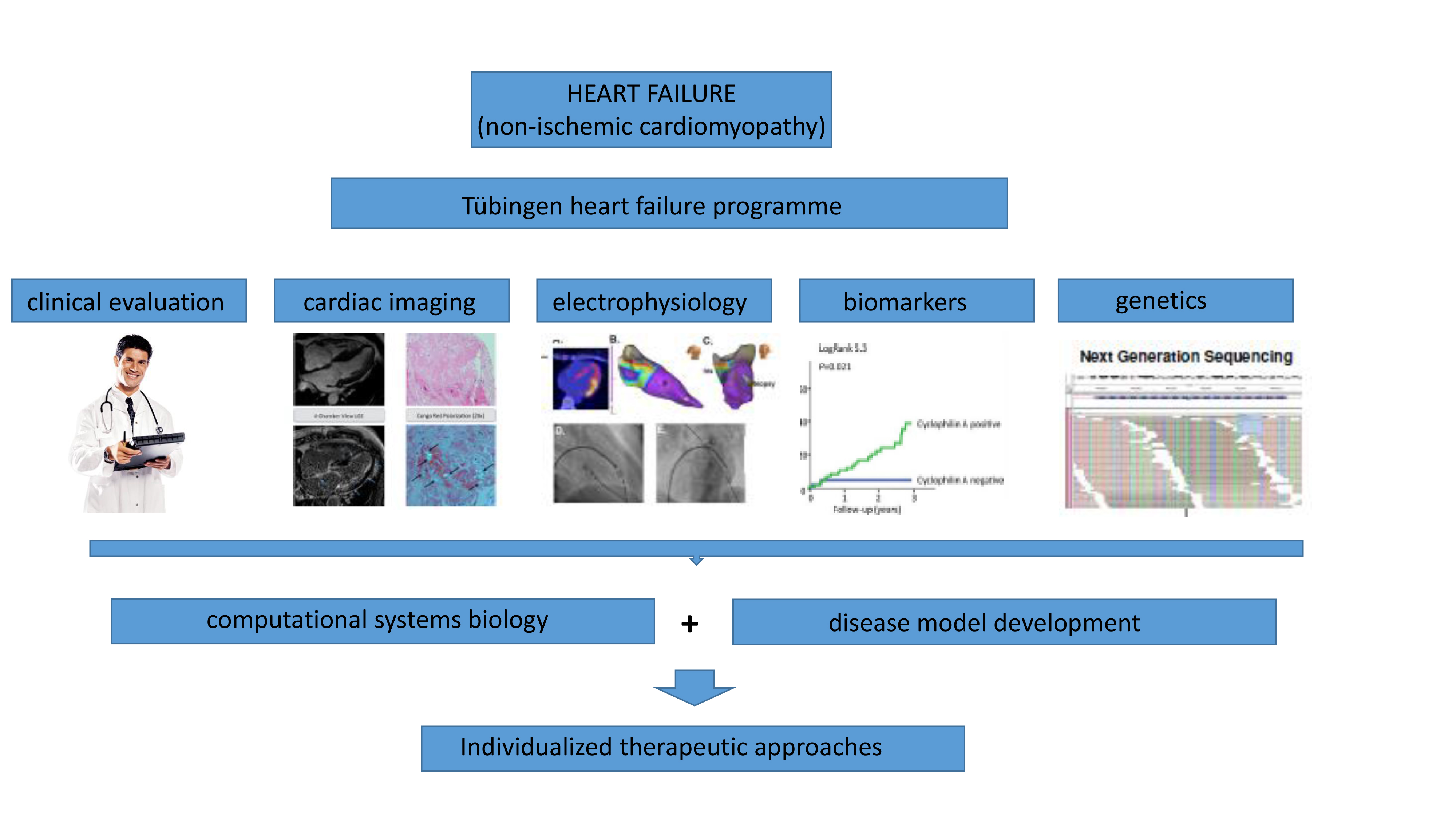

In the last years we have successfully established a heart failure programme at the
Department of Cardiology at the University Hospital of Tübingen. Patients suffering from heart failure receive intensive cardiac imaging (incl. cardiac MRI, PET-CT), heart catheterization, cardiac biospy, electrophysiology, analysis of biomarkers and biosginals as well as the genome. This data will be analyzed in a systems biology approach and predictors for poor clinical outcome or cardiac arrythmias will be established. We've also developped experimental heart failure models (mouse and cell culture models) that serve as a platform to explore novel, individualized therapeutic approaches.
- 1. Analysis of the miRNome, transcritome and genome of heart failure samples
We have established a large biobank of more than 180 transmural heart samples from patients that underwent left-ventricular assist device implantation. We will perform DNA sequencing screen, miRNA and mRNA profiling for all available tissue samples. This unique dataset will be a valuable contribution to understand complex networks in heart failure and to compare Interactions between genetic variants and transcriptional interplay will be analyzed extensively and will be compared across species.
- 2. Evaluation of networks in a mouse line that mimicks a human DCM mutation
We have previously established a DCM mouse line that mimicks a human titin mutation. Extensive mRNA and miRNA profiling during the development of heart failure revealed a variety of dysregulated geneclusters over time. We will further characterize candidate microRNAs and the proposed target genes, methods like quantitative PCR, westernblotting, co-immunoprecipitation studies or luciferase reporter assays will be performed, including functional analysis by blocking (e.g. anti-miRs) respectively overexpression (miRNA-Mimics) of candidate miRNAs in cell culture models and characterization of their cell signaling pathways.
- 3. Charaterization of iPS cell lines from patients with heart failure
We have successfully established reprogramming of dermal fibroblasts into induced pluripotent stem cells (iPS) derived from a patient that carries a titin-mutation. The iPS cells were differentiated into the cardiac lineage and show a DCM-like phenotype. We will use our iPS cell line to produce transcriptome and miRnome profiles. Down this line we will further characterize candidate microRNAs and the proposed target genes.
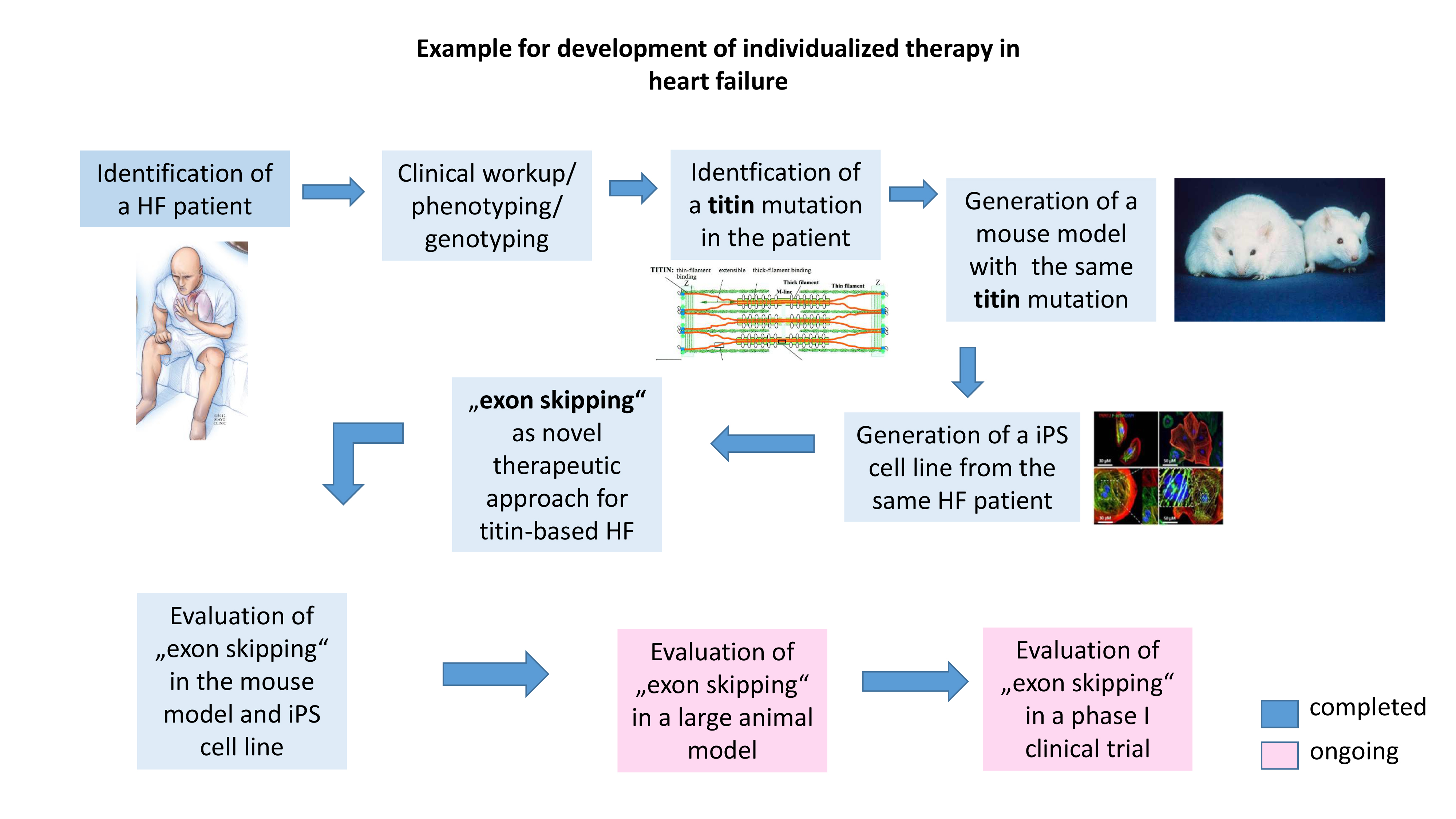

Genetic exploration of a patient suffering from heart failure showed a mutation in the sarcomeric protein Titin. We developed experimental disease models (mouse and iPS cell culture model) expressing exactly the same titin patient mutation. We have established a novel therapeutic approach (so called "exon skipping") and tested it successfully in our disease models. We are currently evaluating this therapy on a large animal model, and then aim to initiate a first clinical trial with diseased patients.



