SP 2 - Try-IBD
Multi-omics data integration via network inference to identify compartment-specific molecular instigators of tryptophan immune-metabolism in intestinal inflammation
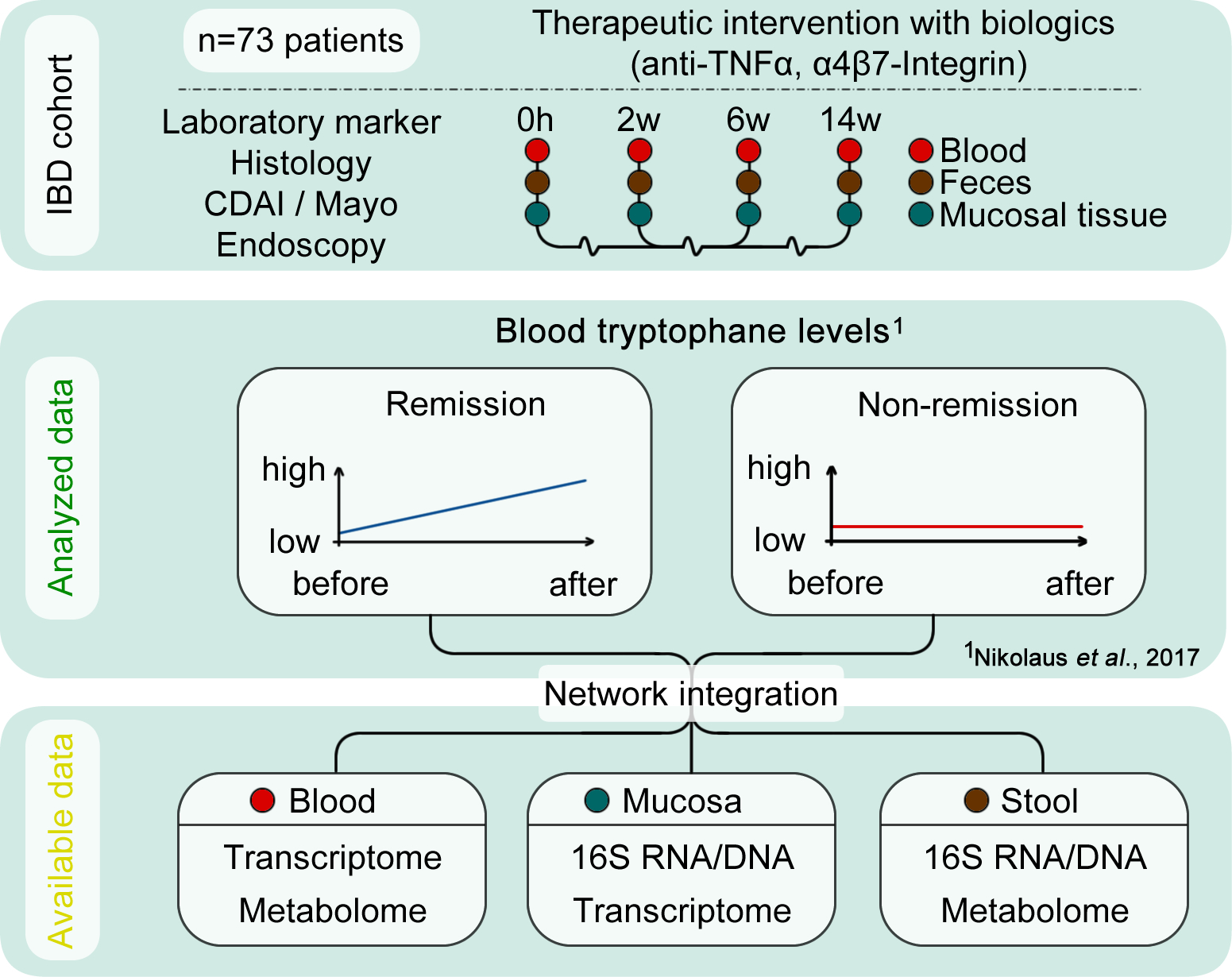
Today, the most common approach to multi-omics data integration is to analyze all 'omic layers' independently and then look for common features. This simple strategy indirectly assumes that all omics levels contribute equally to a biological phenotype, which can lead to misinterpretation of results or overlook the relevant synergistic effects. Moreover, this simple analysis does not capture the complex interaction "cross-omic". There are several applications that take into account all available omic levels to predict clinical outcomes or perform an ontology analysis. Unfortunately, most of these methods require several hundred samples to accurately model the interaction.
Here at the Institute of Compuational Biology, HelmoltzCenter Muenchen, a regression-based Network Inference approach has been developed, which already uses existing knowledge to deduce disease-specific mRNA / miRNA interaction networks. This method has now been extended to work on more than two omic levels, outperforming competing techniques not only in terms of small cohorts (n = 50 patients) but also in terms of accuracy.
In TRY-IBD our goal is to laverage this method for (i) network inference for multiomic data to identify designs for key features, (ii) pathway-centric analysis of multi-omics data to identify key cell functions, and (iii) network validation and extension using in vitro Co-culture models. Finally, the acquired knowledge will be used to (iv) create a machine-learning-based algorithm that classifies molecular subtypes of IBD that influence the therapeutic success of IBD.