SP 1
Telomere maintenance mechanism (TMM) (epi)genomics and transcriptomics
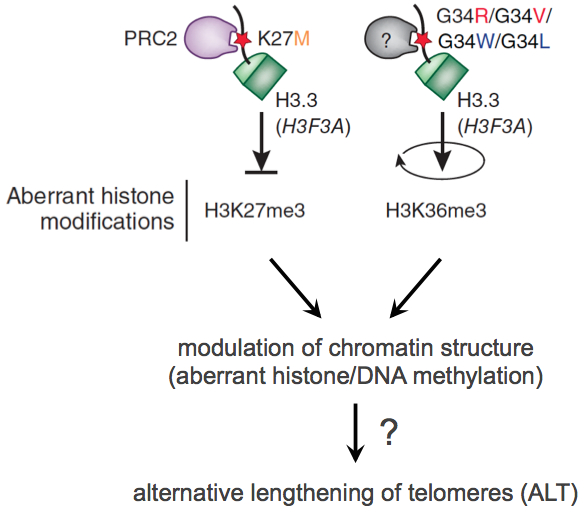
In subproject SP1 we work out a first compilation of key features which are relevant for active Telomere Maintenance Mechanisms (TMM). We analyze existing genome profiling data (gene expression, DNA sequence, DNA methylome) to identify key features of different TMM types in a given tumor sample. Based on the analysis of these features in probes of glioblastoma and prostate cancer we create a list of identified deregulated (epi)genomic TMM features as well as a list of RNAs characteristic for active TMMs. Both lists will be integrated into the TelNet database, which contains also genes that are involved in telomer biology. The TelNet database allows a systematic evaluation of TM features and their integration into TM pathway models or genome-wide mutational analysis of cancer genomes within the CancerTelSys consortium. Additionally, we foster the development of standard protocols for the analysis of high-throughput sequencing data, particularly with regard to noncoding RNAs. Based on their telomere length heterogeneity, a large fraction of the samples have already been classified into putative ALT-negative or ALT-positive (Alternative Lengthening of Telomers, ALT) tumor samples, which allows a first rough grouping. A special focus of SP1 is on the analysis of the ATRX/DAXX/H3.3 pathway, since recent published and unpublished work from the groups Pfister, Plass and Sauter identified these genes as containing driver mutations for pediatric glioblastomas and prostate cancer. These mutations were associated with drastic effects on global DNA methylation pattern, which will also be included as a parameter in the TMM analysis. Certain repair proteins associated with an active TMM are also of particular interest.