SP 6
Epigenetics and transcriptome plasticity in psychiatric diseases
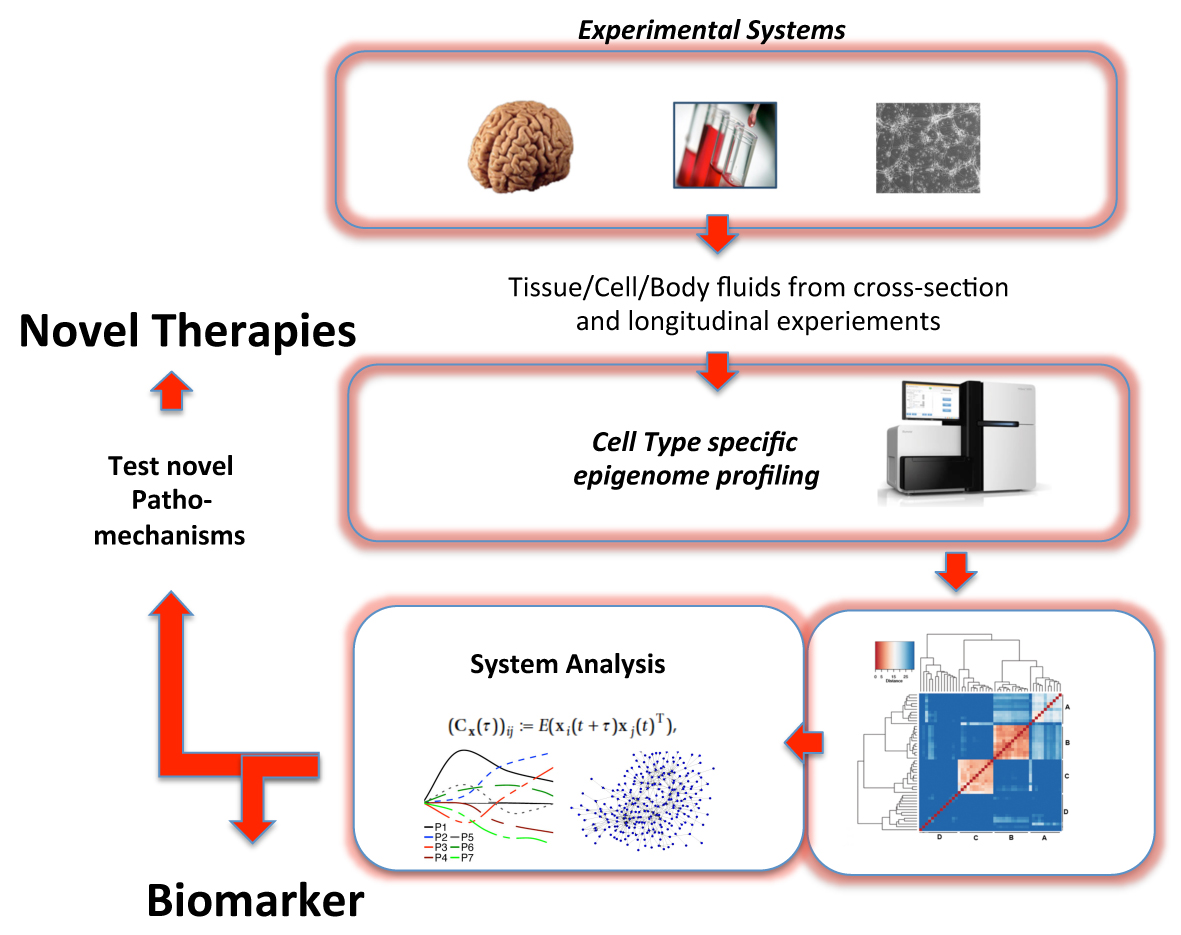
While multiple genetic factors contribute to the etiology of neuropsychiatric disorders, it is now commonly accepted that the presence of a healthy vs. diseased state is critically dependent upon an interaction between genes and environment. Epigenetic mechanisms regulate long-lasting changes in gene-expression in response to environmental stimuli, and the deregulation of epigenetic processes contributes to the pathogenesis of neuropsychiatric disease. Thus, improved understanding of disease associated changes in the epigenetic network will help to identify biomarkers and thus facilitate individualized treatment. The aim of SP6 is to generate epigenetic profiles in combination with the transcriptome for the purposes of systems analysis using next-generation sequencing technology (NGS). More specifically, we will perform genome-wide level analyses of DNA-methylation, DNA-hyrdoxymethylation, and histone-methylation in a cell type specific manner in different brain regions, using tissue from well characterized schizophrenia patients and controls. The data will be combined with the analysis of the entire coding and non-coding RNAome (including micro RNAome) using RNA-sequencing. Similar analyses will be performed in blood samples and in iPS cells available to the Consortium, since there is evidence that peripheral epigenetic bio-signatures reflect changes in centrally relevant mechanisms. The integration of our data (in cooperation with SP1) with the findings of the other SPs (SP3, SP4, SP5, SP7 and SP8) will allow us to identify disease signatures that point to novel pathomechanisms and will help to develop individualized therapies.
Keywords: Epigenetics, non-coding RNA, psychiatric diseases



