SP 3
Central Resource II: Transcriptomics platform
The transcriptome is the first stage at which genetic variability affects neurobiology. Without comprehensive databases on human gene expression in alcoholic patients the attempted goals of this consortium cannot be achieved, because neither genetic variation nor brain activation can be understood without considering their consequences onto the molecular machinery. The objective of this SP3 is to provide a transcriptome database from human brain and inducible pluripotent stem cells (iPSCs) of alcoholics and control subjects as well as from corroborative animal models.
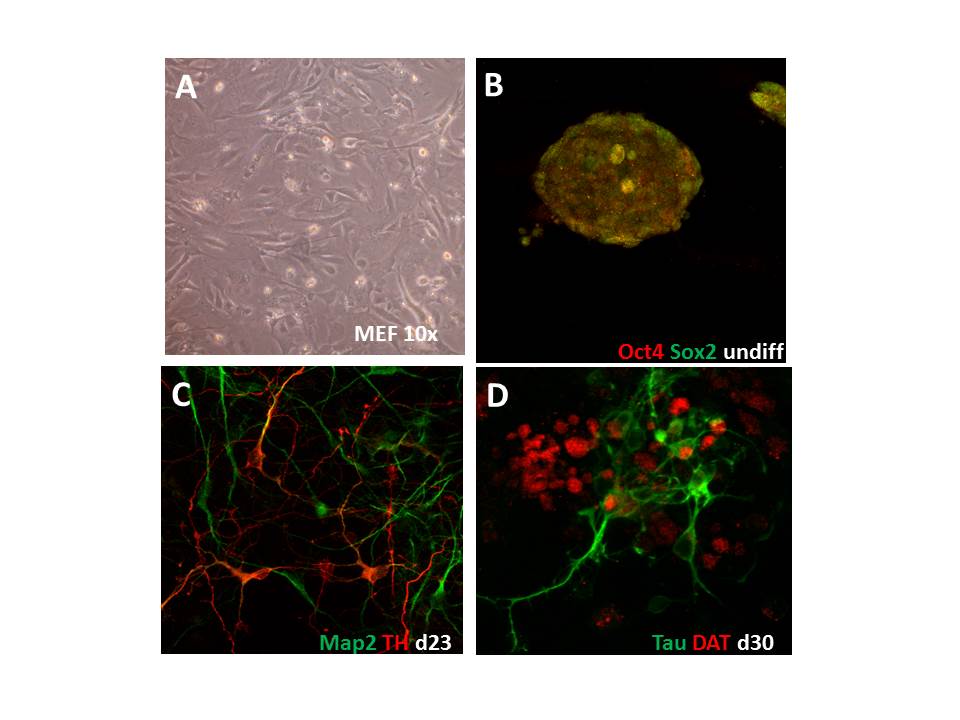
This database will provide critical information for identifying and validating single nucleotide polymorphisms (SNPs) and DNA methylation sites obtained in SP2, to identify potential targets for functional validation in SP9 and 10, and to deliver gene lists from various experiments and sources to the convergent data analysis and statistics SP6. Human expression data will be obtained from postmortem brains from reward-related brain regions (i.e. prefrontal cortex, striatum). This sample set will be used to test SNPs obtained from SP2 for effects on mRNA (qPCR) and protein level. Human expression data will be also obtained from neurons of patients with high genetic load generated by iPSC technology. Expression data from various animal models of alcoholism (including the DSM-IV/V-like rat model described in SP5) are already available from previous NGFN and EU-funded projects. In these human and animal-derived brain tissues we will investigate convergent mRNA and microRNA profiles, the former to integrate with genetic association and DNA methylation studies (SP2), the latter because they are master regulators of gene expression networks. The combined data from deceased alcoholics, treatment seeking patients and from carefully selected animal models will provide the most comprehensive expression database on alcoholism so far, which will be a unique resource for the consortium and ultimately for the scientific community.



