SP 10
Data management and multi-scale data analysis
The e:KID project generates a spectrum of different data types including clinical data, gene expression data, cytokine data, epigenetics data, metabolomics data as well as data derived from HLA antibody measurements and viral load measurements. In order to allow comprehensive data analysis all the different data types have to be integrated into one common resource. For achieving this goal, MicroDiscovery GmbH can resort to long established methods and experiences [BioMiner: Paving the Way for Personalized Medicine. (PMID: 26005322)]. In the e:KID project we create and manage an integrative database resource for all the data generated in the consortium. The database is updated on a regular basis and will be provided to all project partners.
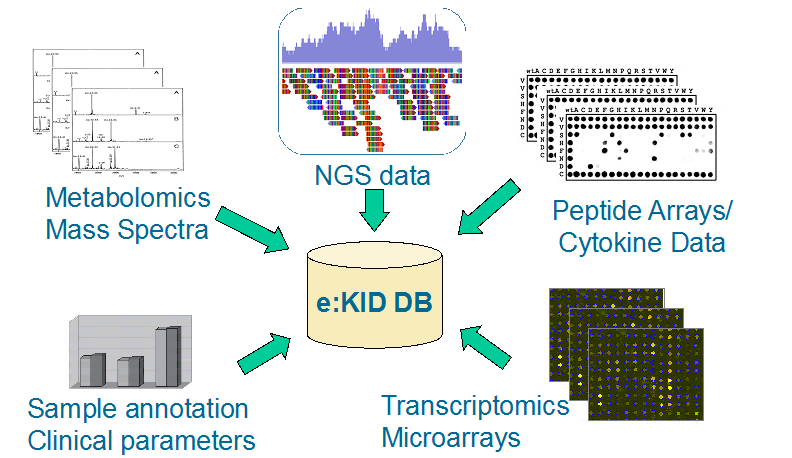
Integration of multiple data sources into the e:KID database
Besides data management, MicroDiscovery is contributing to data analysis tasks. The data generated in the project is not only cross-omics but also time-resolved, heterogeneous and high-dimensional. To discover hidden connections between multi-scale markers multi variate statistics and time series analyses are applied to the data. These analyses allow to identify biomarkers associated with a positive long-term graft outcome and will help to develop personalized treatments for patients requiring kidney transplants.
Methods:
• Cross-OMICs data integration and data management
• Relational database management systems
• Quality control, data normalization, multivariate imputation
• Machine learning, multivariate classification and feature selection
• Identification of pathways and biologically relevant networks



