SP 9
Epigenetic biomarkers for risk assessment, prognosis, and prediction of post-transplant course upon kidney transplantation
It is necessary to adjust permanently the therapy of kidney transplant recipients with immunosuppressive drugs to avoid the risk of graft rejection and side effects of therapy, like an increased liability to infections and therapy related tumor formation. Today’s available diagnostic methods for post-transplant complications are partially invasive (such as transplant biopsy) and don’t allow the sufficiently early prediction of complications to adjust the immunosuppressive therapy to avoid complication. For this reason non-invasive or low-invasive predictive diagnostic tests are preferable.
The aim of this study is the identification of quantitative changes of clinical relevant immune cells like effector T cells (Teff), regulatory T cells (Treg), natural killer cells, monocytes, Th17 and B cells in both whole blood and urine samples, for early detection of immunological complications post transplantationem. Therefore, the ratio of Teff to Treg could be a possible indicator. The suitable clinical quantification of kidney infiltrating immune cells as well as the specific Teff/Treg-infiltration can only be ensured by epigenetics. The epigenetic analysis is based on the representation of immune cell type specific DNA methylation patterns (epigenetic markers) by real-time PCR. Thus, defined immune cell populations can be quantified with high-sensitivity and precision. With this valid method the discrimination of Tregs for instance, which shows a complete demethylated region in the Foxp3-gene in contrast to complete methylation in this region in the other cell types, is possible. Epiontis Ltd. has comprehensive expertise in development of epigenetic assays, including marker discovery, to identify an acute graft rejection to individually adjust an immunosuppressive therapy.
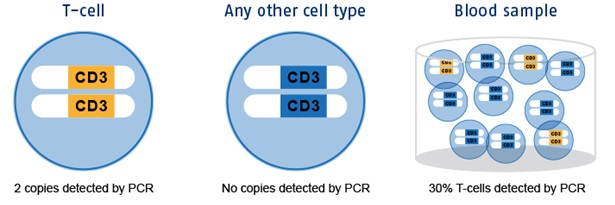
The blue circles are T cells respectively any other immune cell in blood. These cells contain chromosomes with the CD3 gene region. Yellow regions represent the non-methylated (epigenetic active) variant in T cells, the blue ones are methylated (epigenetic non-active) in any other immune cell. With an immunomonitoring-assay only the epigenetic active alleles can be amplified and detected by PCR. Non-T cells are not detected. The number of epigenetically active CD3 gene copies directly translates into the number of T cells in the sample. Parallel measurements of epigenetic reference systems, e.g., housekeeping gene GAPDH or specific plasmid standards, allow for total cell number determination.
Keywords: Treg, regulatory T cells, Kidney rejection, epigenetic markers



