SP 5
In situ proteome analysis of gastric cancer
The deregulation and re-programming of signalling pathways and networks underlie tumour development and acquired resistance to therapy. Two proteins known to be over-exressed in gastric cancer, Human Epidermal Growth Factor Receptor 2 (HER2) and Epidermal Growth Factor Receptor (EGFR), stimulate tumour cell growth and survival. Clinical trials of novel drugs targeting these receptors or downstream signalling have shown mixed patient responses. This subproject will conduct in-situ molecular analysis to examine mechanisms of response or resistance to EGFR and HER2-based therapies. Matrix-Assisted Laser Desorption/Ionisation Imaging Mass Spectrometry (MALDI-MSI) is a powerful tool for investigating the distribution of molecules in situ. Unlike classical histology, MALDI-MSI is able to detect numerous molecules in a single sample. Additionally, MALDI-MSI is able to maintain the spatial distribution of molecules – information that is lost in liquid-based mass spectrometry techniques, and which is of particular importance given the increasing evidence that gastric tumours are molecularly heterogenous. MALDI-MSI will be used to identify and evaluate biomarker candidates in gastric cancer, and expand the body of knowledge on HER2/EGFR signalling pathways.
Using MALDI-MSI, SP5 will conduct measurements geared towards the discovery of biomarkers, increasing knowledge of networks, and protein, drug and metabolite signatures associated with drug response. These findings would be validated in a clinical setting for patients.
This subproject is organised into three work packages addressing specific aims while providing critical information for consortium partners.
1. Clinical evaluation of proteomic biomarker candidates
We have previously generated MALDI-MSI protein expression profiles from gastric cancer patients that indicated differential expression of proteins between individuals related to HER2 and EGFR status, tumour progression, and survival. This data will be provided to SP2, SP3, and SP4 to further identify expression profiles and nominate proteins to indicate responding/non-responding tumours.
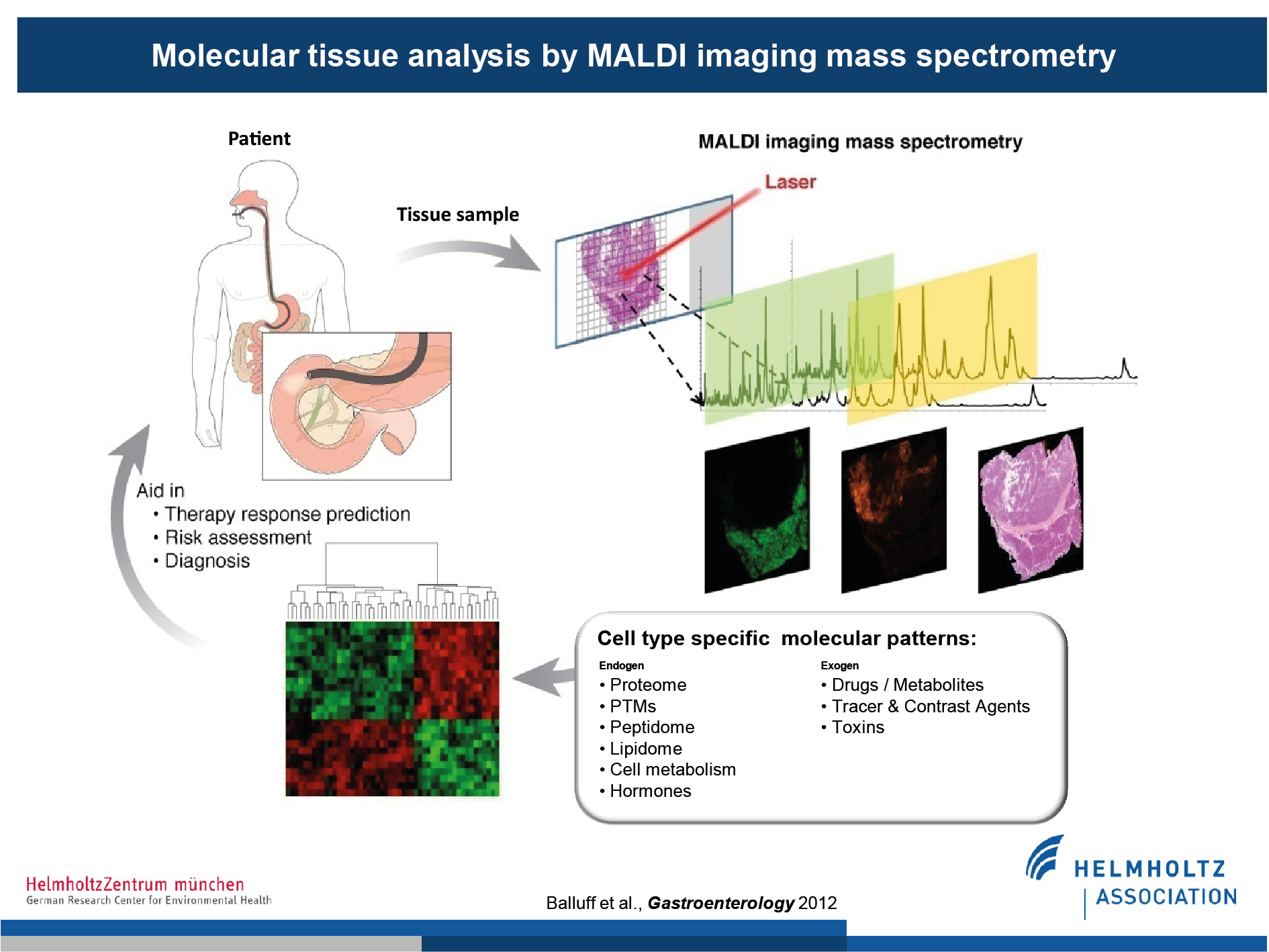
Patient samples, such as gastric biopsies, can be measured via MALDI-MSI. Measurements are then co-registered to corresponding histological sections, allowing the identification of spatially resolved molecular signatures. These molecules are derived from a wide variety of classes, e.g. proteins, lipids, drug and drug metabolites, and can be used to address clinically relevant questions such as predication of therapy response or resistance.
(Figure modified from Balluff et al., Gastroenterology, 2012)
2. HER2 and EGFR network Analysis.
In order to expand and refine the HER2 and EGFR signaling pathways, we will measure molecular expression from drug-responding/non-responding cell lines generated by SP1. This data will identify new proteins for network modelling (SP3) as well as providing information about cross-talk into cancer-relevant pathways.
3. Proteomic signatures of patient response/non-response to HER2 and EGFR-based therapies.
Samples from patient cohorts of clinical trials of HER2 and EGFR therapies from SP6 will be measured with MALDI-MSI and directly compared and integrated with data from cell lines by SP2, SP3, and SP4.
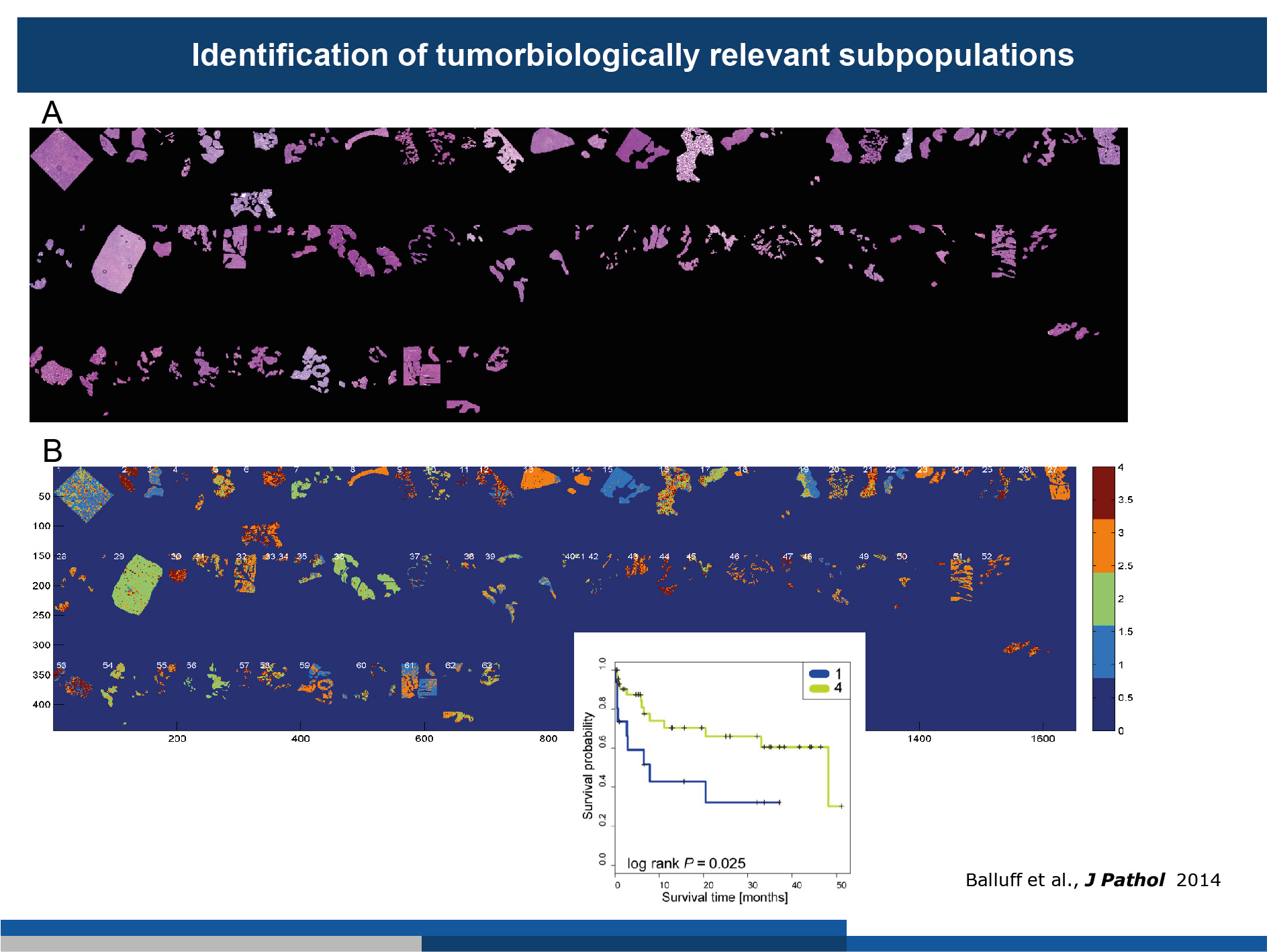
(A) 63 gastric cancer patient samples were measured by MALDI-MSI. The segmented tumour cells are shown as haematoxylin and eosin stainings.
(B) Agreement analysis revealed substantial inter- and intra-tumour heterogeneity. By correlation of the tumour subpopulations with the survival data, we were able to determine those tumour subpopulations which were linked to patient survival (inset).
(Figure modified from Balluff et al., J Pathology, 2014)



