SYSMED-NB
Systems Medicine for Neuroblastoma
The major challenge impeding optimal treatment outcomes for cancer patients arises from the complex nature of the disease. The rise of high-throughput technologies and computational strategies to comprehensively assess tumor DNA, RNA and proteins has fueled efforts to tailor medical care for individual cancer patients, including the devastating childhood tumor, neuro-blastoma (NB). Our previous efforts in molecular profiling of NBs have provided potential targets for novel therapies and a wealth of data for optimized risk stratification. Managing the data flood generated and converting this to the knowledge required to improve patient outcomes is chal-lenging. We propose a comprehensive systems medicine approach to NB employing innovative data management strategies and systems biology modeling with its holistic approach to intricate interactions of networks controlling biological functions. This cross-disciplinary concept offers the tools to deconvolute the plethora of molecular pathogenetic factors to core principles with therapeutic relevance. The central research goal is to identify and understand core signaling networks controlling biological behavior of highly aggressive, metastasized NBs focusing on molecules interacting with the major NB key player, MYCN. Two synergistic research pipelines (A) systematically explore existing and complementary “omics” datasets via innovative mathe-matical modeling approaches to move towards efficient biomarker use and target identification within clinical trials and (B) operate a systems biology-driven discovery and validation engine to investigate key players in NB, identify nodes vulnerable to therapeutic targeting and develop individualized treatment strategies. The deeper understanding of signaling networks driving NB pathogenesis will be used to create the most effective targeting strategies supported by precise selection of appropriate patient subgroups according to individual risk and molecular biomarker profile.
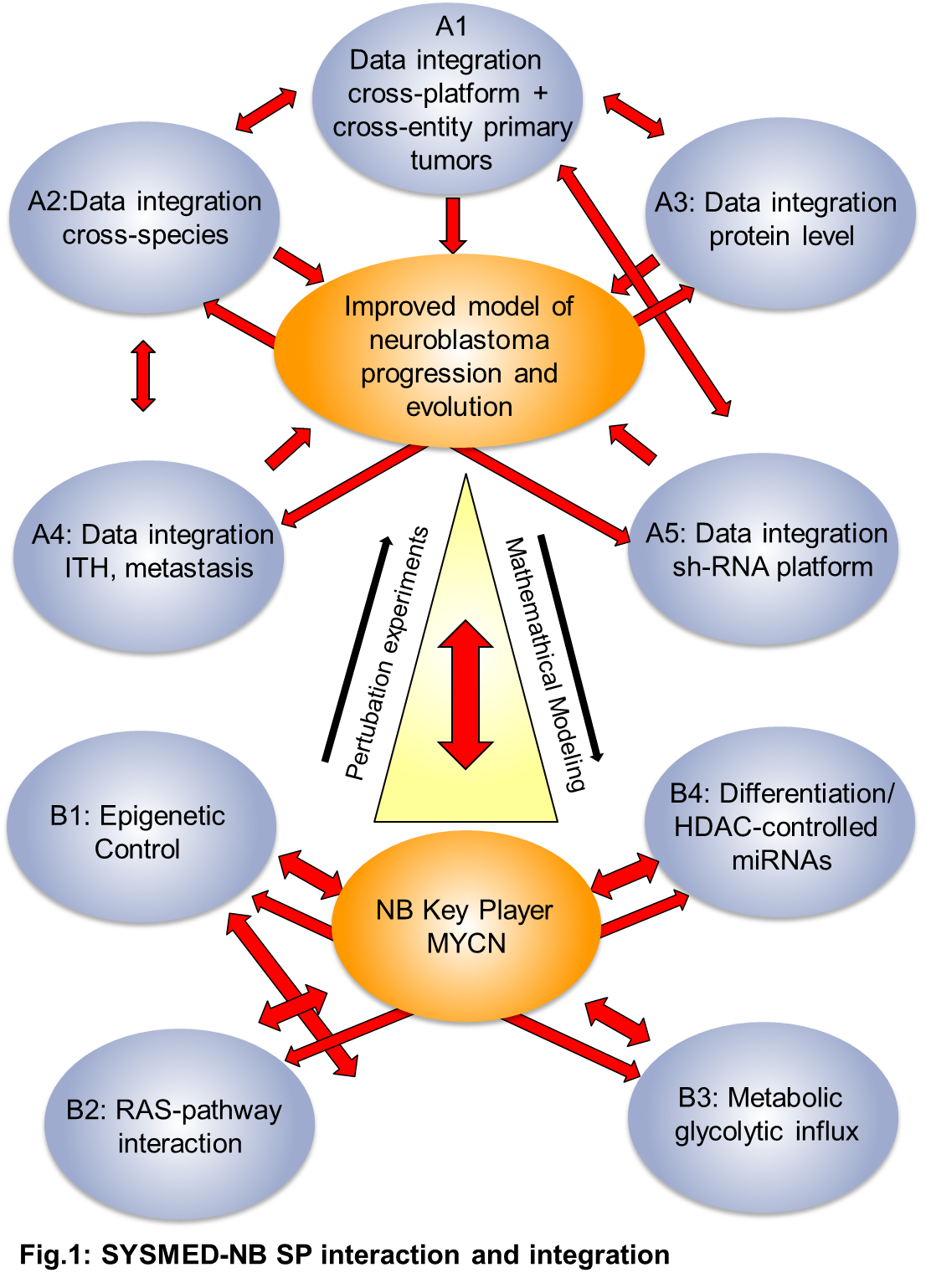
Subprojects in SYSMED-NB
- SP A2 Mouse modeling and crossspecies analysis to optimize identification and targeting of MYCN co-drivers in NB
- SP A5 Translating genomic information to therapeutic targets for neuroblastoma using systematic loss-of-function screening
- SP B2 Targeting the RAS pathway in high-risk neuroblastomas
- SP B3 Modeling primary sugar metabolism in neuroblastoma to identify central nodes for therapeutic intervention
- SP B4 Network analysis of HDAC-controlled microRNAs in neuroblastoma



