SP 3
Identification of disease-associated gene regulatory networks
In the last 10 years, many genome-wide associating studies (GWAS) have been conducted on most common non-infectious diseases. The aim was to find the genetic variants that confer a higher risk to develop the disease under investigation. First, this could allow us to predict which patients are particularly at risk and to propose measures to delay or prevent the disease. But second, studying the genetic variants offers the fascinating opportunity to identify causes and mechanisms underlying the disease. To find the high-risk variants, in GWAS the genomes of patients with the disease are compared with genomes of a control group of healthy people. Statistical and computational methods then predict the disease-associated variants.
Our eAtheroSysMed consortium investigates the origins and causes of atherosclerotic diseases such as stroke and coronary artery disease (CAD), in which the arteries are obstructed by slowly accumulating plaques on the inner walls of blood vessels.
Many genetic risk variants have been found in recent GWAS. However, often the apparently affected genes cannot be interpreted functionally. Recent evidence from genome-wide functional characterizations of the human genome points to a likely reason for this difficulty: In the past, we have assumed that the gene nearest in sequence to the genetic variant will be affected by this variant. However, it is likely that often the genetic variant does not affact the nearest gene but modifies the gene activity of a more distant gene.
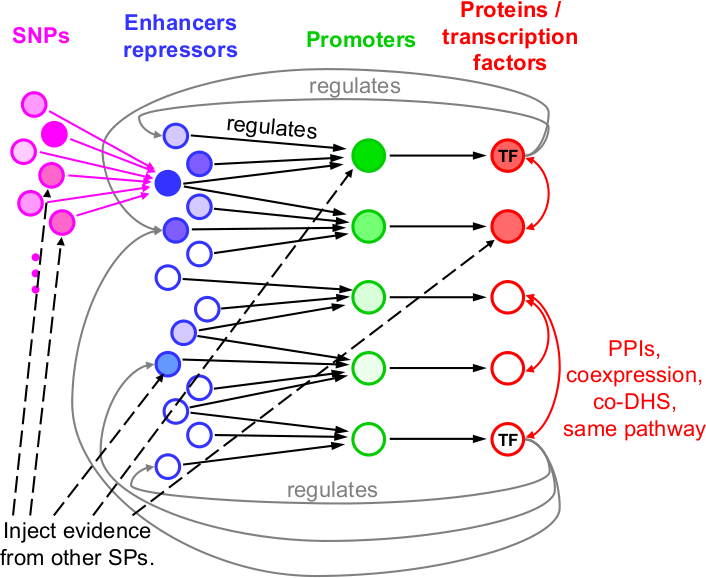
In this subproject our first goal is therefore to develop methods to predict the target genes of regulatory genetic variants. Our second goal is to derive the regulatory networks that are affected by the many genetic variants discovered in the genome-wide association studies on coronary artery disease and stroke. We will develop novel statistical and computational approaches to integrate information from genetic variants, disease type, and gene expression data with many types of public data that characterizes the functional activities in the human genome. Our models will effectively allow us to aggregate signals for the association of genes with atherosclerosis over numerous genetic variants affecting the same atherosclerotic pathways. In this way we hope to discover atherosclerosis-associated regulatory networks that are deregulated by the genetic variants identified in GWAS and thus contribute to the increase of disease risk.
Keywords: Genome-wide association studies, atherosclerosis, CAD, stroke, computational biology, systems medicine, network analysis, enhancers, regulatory SNPs, regulatory networks



